RBP Type
Non-canonical_RBPs
Diseases
SARS-COV-2
Drug
Main interacting RNAs
N.A.
Moonlighting functions
Metabolic enzyme
Localizations
N.A.
BulkPerturb-seq
Ensembl ID ENSG00000206172 Gene ID
3039 Accession
4823
Symbol
HBA1
Alias
HBH;ECYT7;HBA-T3;METHBA
Full Name
hemoglobin subunit alpha 1
Status
Confidence
Length
843 bases
Strand
Plus strand
Position
16 : 176680 - 177522
RNA binding domain
N.A.
Summary
The human alpha globin gene cluster located on chromosome 16 spans about 30 kb and includes seven loci: 5'- zeta - pseudozeta - mu - pseudoalpha-1 - alpha-2 - alpha-1 - theta - 3'. The alpha-2 (HBA2) and alpha-1 (HBA1) coding sequences are identical. These genes differ slightly over the 5' untranslated regions and the introns, but they differ significantly over the 3' untranslated regions. Two alpha chains plus two beta chains constitute HbA, which in normal adult life comprises about 97% of the total hemoglobin; alpha chains combine with delta chains to constitute HbA-2, which with HbF (fetal hemoglobin) makes up the remaining 3% of adult hemoglobin. Alpha thalassemias result from deletions of each of the alpha genes as well as deletions of both HBA2 and HBA1; some nondeletion alpha thalassemias have also been reported. [provided by RefSeq, Jul 2008]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
Name
Transcript ID
bp
Protein
Translation ID
HBA1-201
ENST00000320868
577
142aa
ENSP00000322421
HBA1-203
ENST00000472694
674
No protein
-
HBA1-202
ENST00000397797
504
110aa
ENSP00000380899
HBA1-204
ENST00000487791
410
No protein
-
Pathway ID
Pathway Name
Source
ensgID
Trait
pValue
Pubmed ID
76598 Erythrocyte Indices 3E-9 20139978 76599 Hemoglobins 3E-9 20139978
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
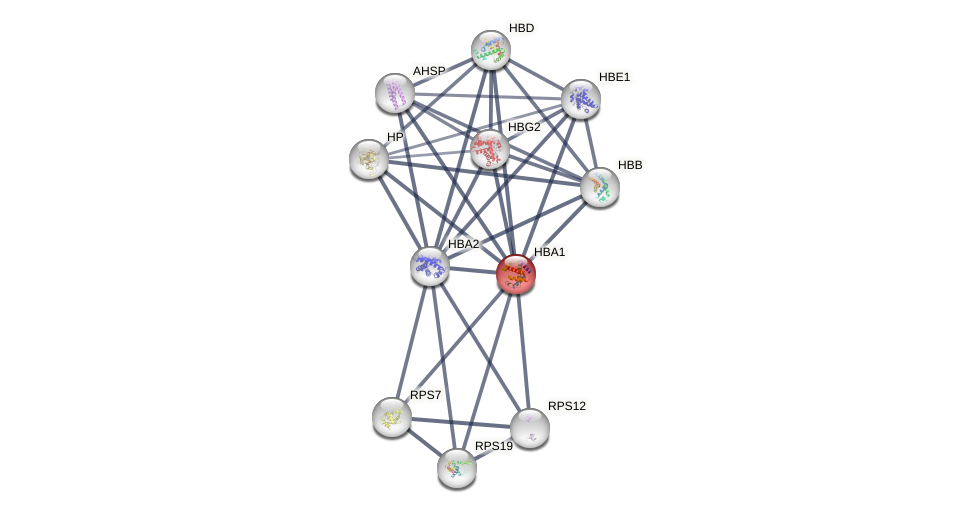
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0004601 contributes_to peroxidase activity 2 IBA,IDA Homo_sapiens(9606) Function GO:0005344 enables oxygen carrier activity 1 IBA Homo_sapiens(9606) Function GO:0005506 enables iron ion binding 1 IEA Homo_sapiens(9606) Function GO:0005515 enables protein binding 1 IPI Homo_sapiens(9606) Function GO:0005576 located_in extracellular region 1 TAS Homo_sapiens(9606) Component GO:0005615 located_in extracellular space 1 IDA Homo_sapiens(9606) Component GO:0005829 located_in cytosol 1 TAS Homo_sapiens(9606) Component GO:0005833 part_of hemoglobin complex 4 IBA,IDA,IPI,TAS Homo_sapiens(9606) Component GO:0015670 involved_in carbon dioxide transport 1 NAS Homo_sapiens(9606) Process GO:0015671 involved_in oxygen transport 4 IDA,IMP,NAS,TAS Homo_sapiens(9606) Process GO:0016020 located_in membrane 1 HDA Homo_sapiens(9606) Component GO:0019825 enables oxygen binding 1 IBA Homo_sapiens(9606) Function GO:0020037 enables heme binding 1 IBA Homo_sapiens(9606) Function GO:0030185 involved_in nitric oxide transport 1 IDA Homo_sapiens(9606) Process GO:0031720 contributes_to haptoglobin binding 2 IBA,IDA Homo_sapiens(9606) Function GO:0031838 part_of haptoglobin-hemoglobin complex 2 IBA,IDA Homo_sapiens(9606) Component GO:0042542 involved_in response to hydrogen peroxide 1 IDA Homo_sapiens(9606) Process GO:0042744 involved_in hydrogen peroxide catabolic process 2 IBA,IDA Homo_sapiens(9606) Process GO:0043177 enables organic acid binding 1 IBA Homo_sapiens(9606) Function GO:0070062 located_in extracellular exosome 1 HDA Homo_sapiens(9606) Component GO:0071682 located_in endocytic vesicle lumen 1 TAS Homo_sapiens(9606) Component GO:0072562 located_in blood microparticle 1 HDA Homo_sapiens(9606) Component GO:0098869 involved_in cellular oxidant detoxification 1 IEA Homo_sapiens(9606) Process
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.