Welcome to
RBP World!
Introduction
RBPWorld is an updated version of EuRBPDB, specifically designed to unveil the functions and disease associations of RNA-binding proteins (RBPs) with heightened efficacy. Within RBPWorld, an expansive collection of 1,393,686 RBPs across 445 species, including 3,303 human RBPs (hRBPs). Through RBPWorld, users can easily access information about the downstream regulatory networks, RNA partners, disease associations, and known targeted drugs associated with diverse hRBPs. Use the Google Chrome browser for best visualization quality.
User manuals
To search for a gene of interest, enter a gene symbol/alias/RefSeq id/Ensembl id/OMMI id/HGNC id/Uniprot id/Entrz Gene id in the search box and click the “Go RBP!” button.
Basic information subpage
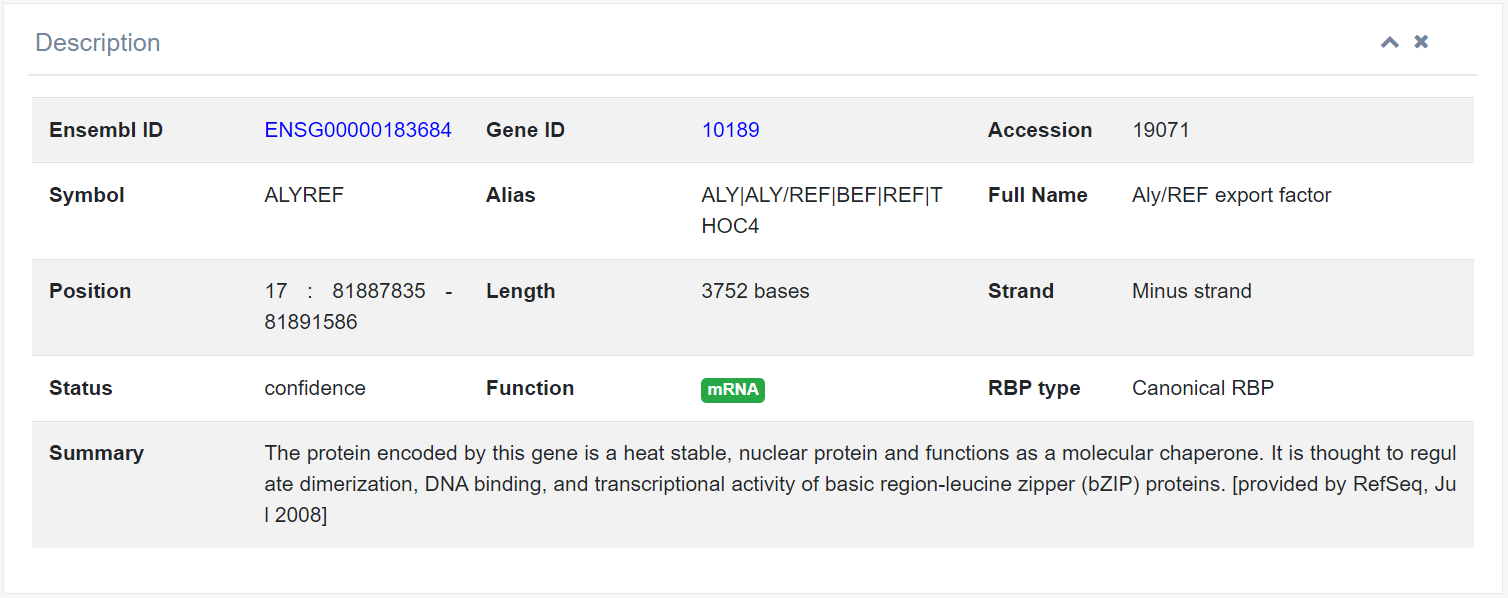
Taking the "ALYREF " protein as an example, users can explore the details of this RBP.
1. Information
This section provides Ensembl ID, Gene ID, Gene Symbol, Alias, Full Name, Gene Type, Strand, Length, Position, and Transcripts information from the Ensembl or GeneCards database.
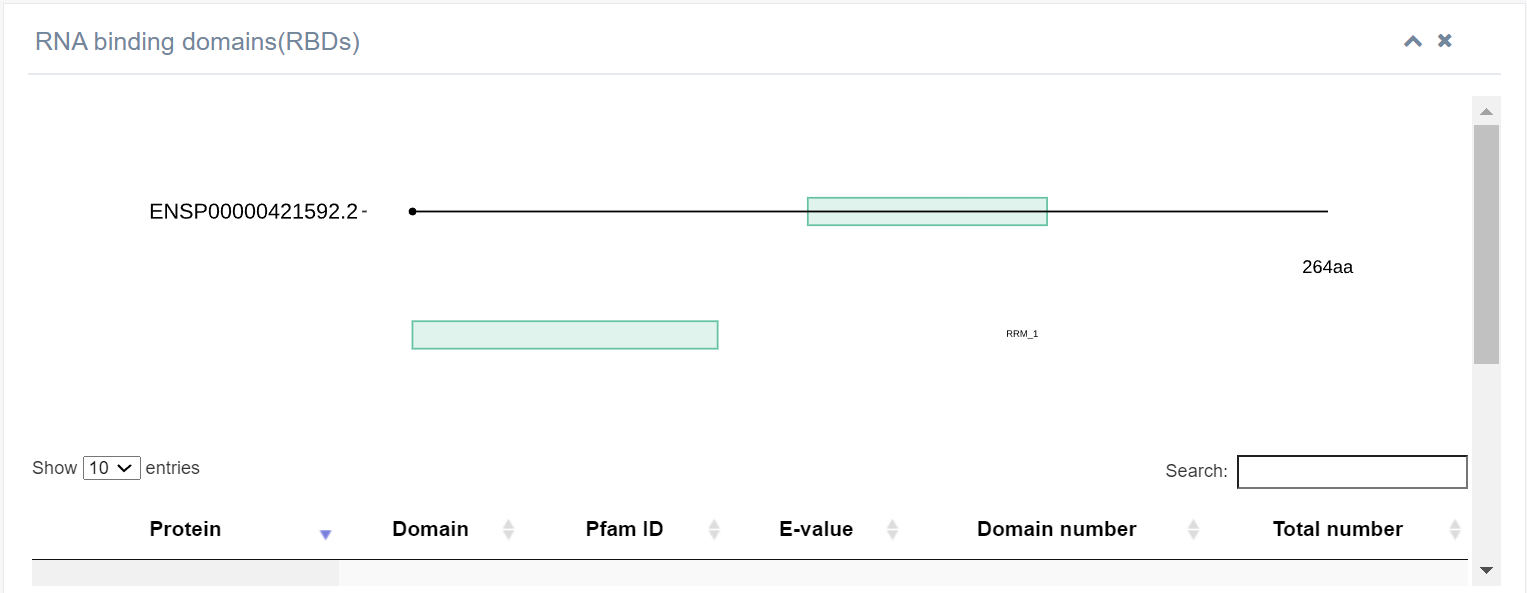
2. RNA binding domains (RBDs)
The figure displays details of the RNA-binding domain present in the ALYREF protein, discovered using hmmsearcher.
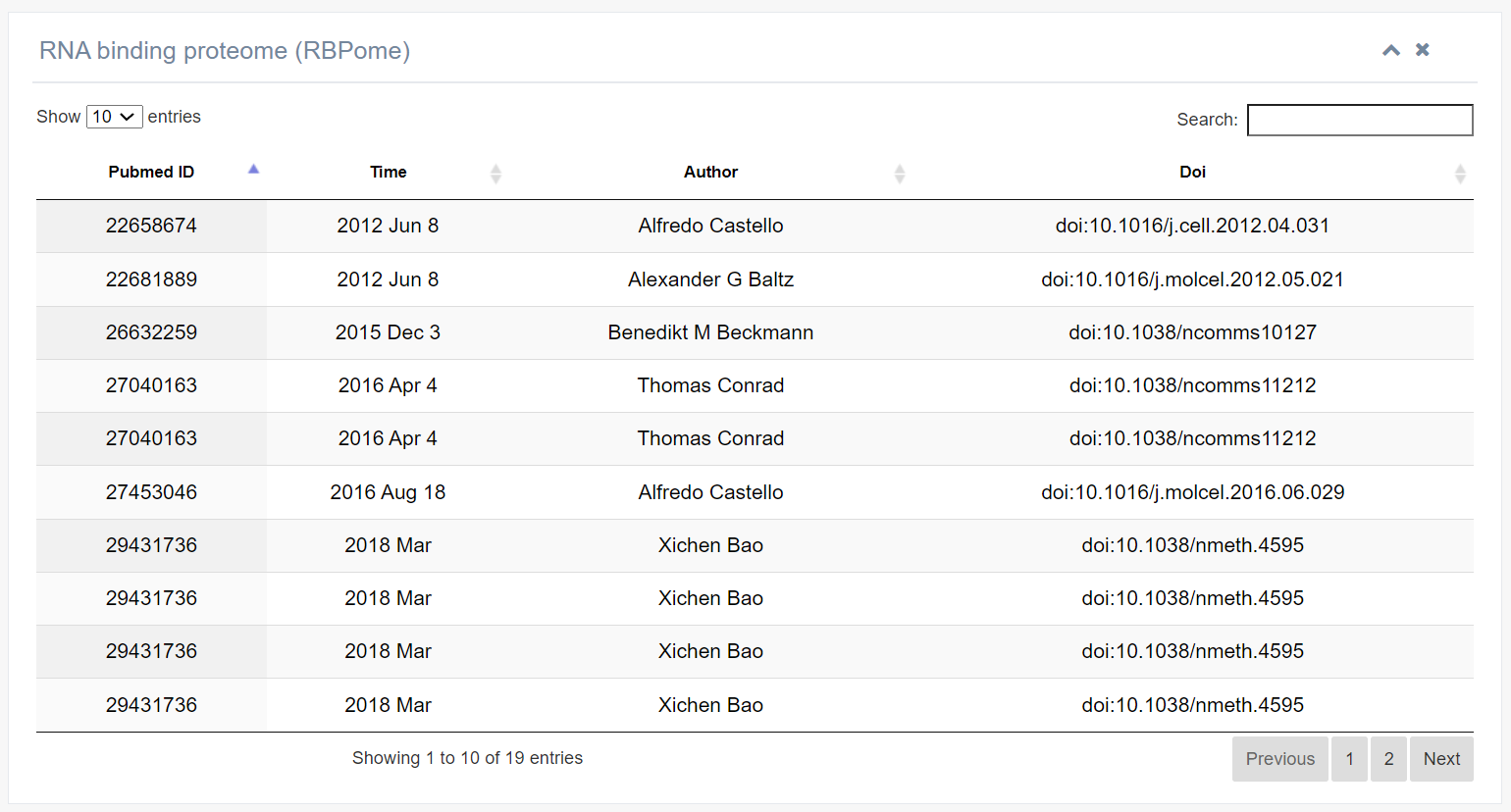
3. RNA binding proteome (RBPome)
This table summarizes recent published literature on RNA-binding proteomics involving the ALYREF protein.
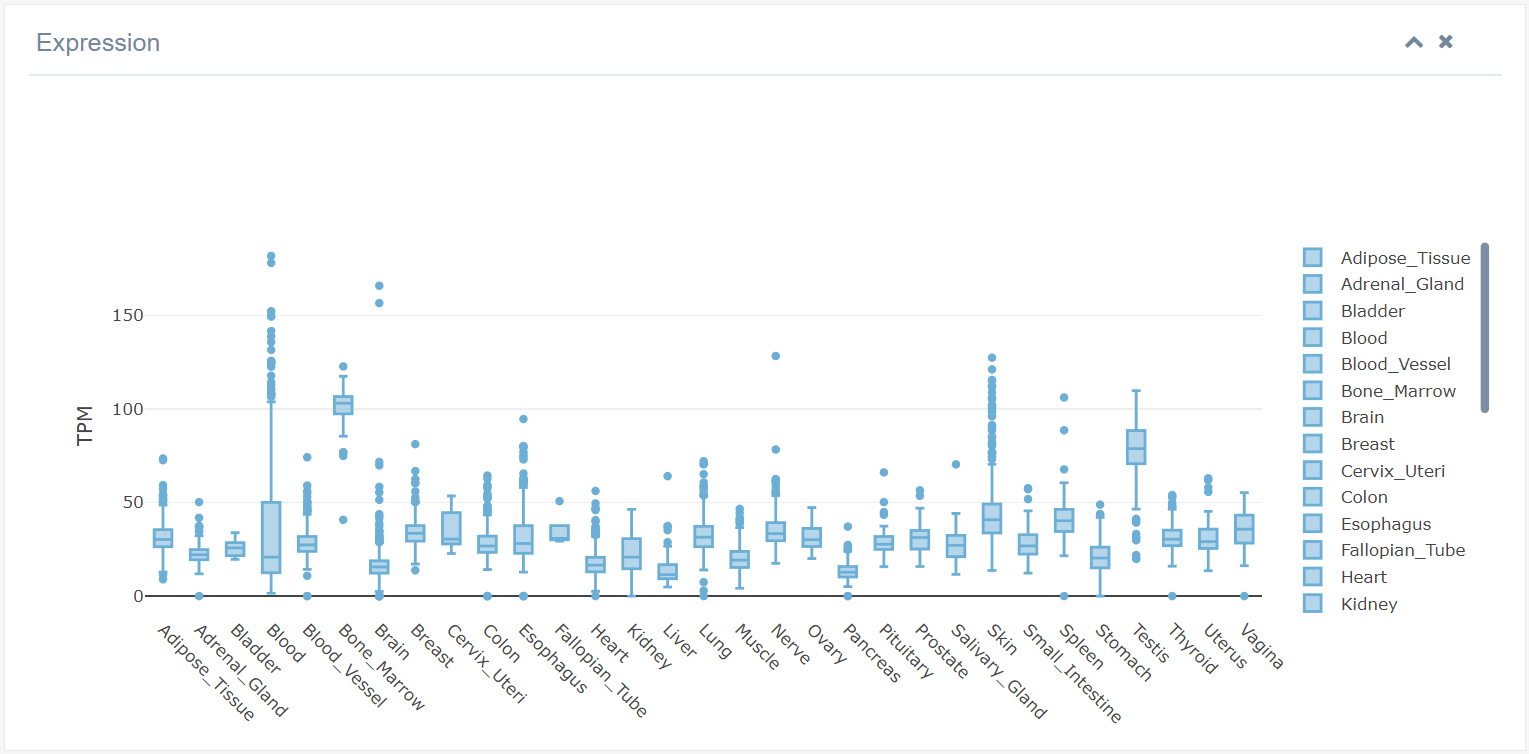
4. Expression
Expression levels of RBPs in different tissues were obtained from public databases like GTEx. EuRBPDB currently contains abundant RBP expression information for animals, plants, and fungi. Boxplots depict the expression level of each RBP, allowing users to add or remove samples by selecting their names in the right panel.
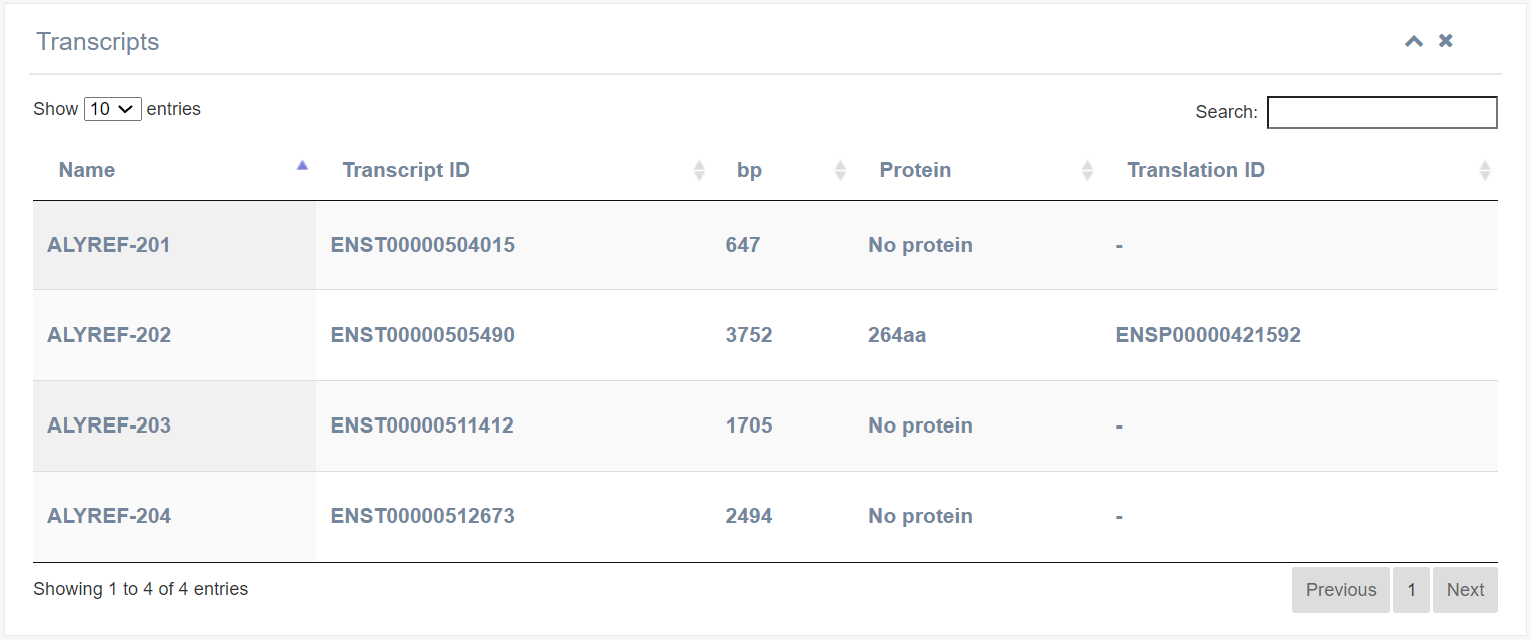
5. Transcripts
This section presents all isoforms of a RBP gene. Users can obtain Ensembl transcript ID, Name, length RefSeq ID, Ensembl protein ID, protein length, and UniportKB ID for each isoform.
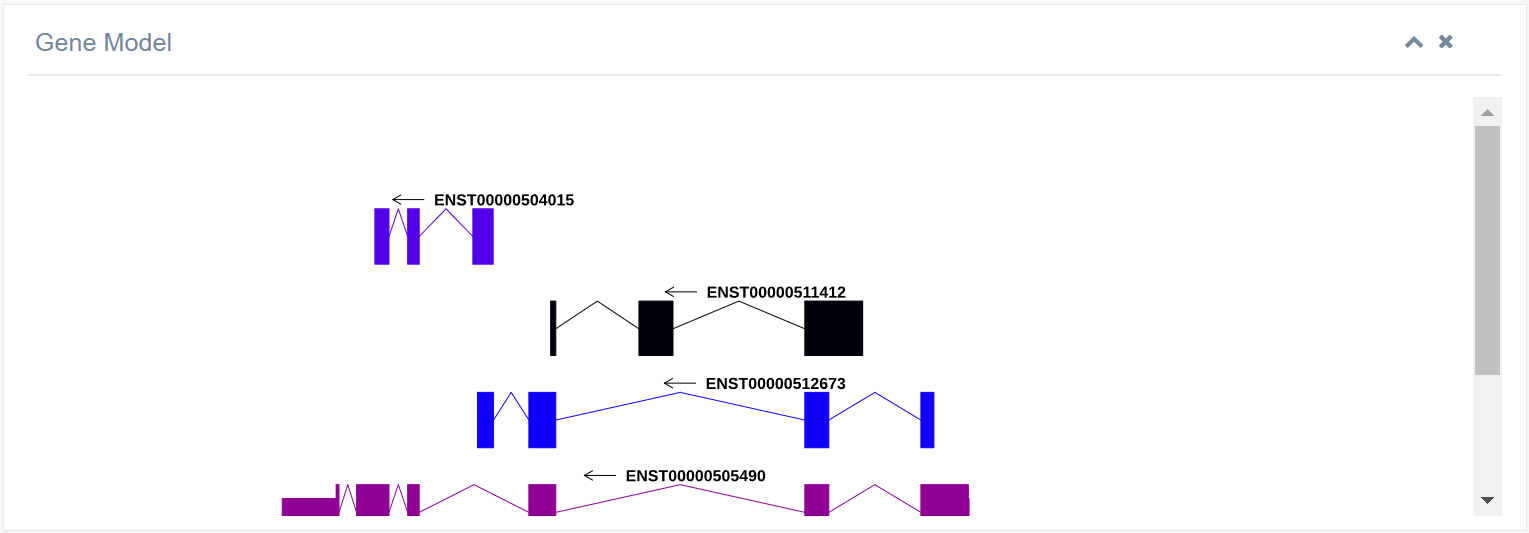
6. Gene Model
This section shows the distribution of CDS, UTR, and intron regions of a gene on the chromosome, based on Ensembl gtf files. Clicking the link in the lower figure can provide a high gene corner model.
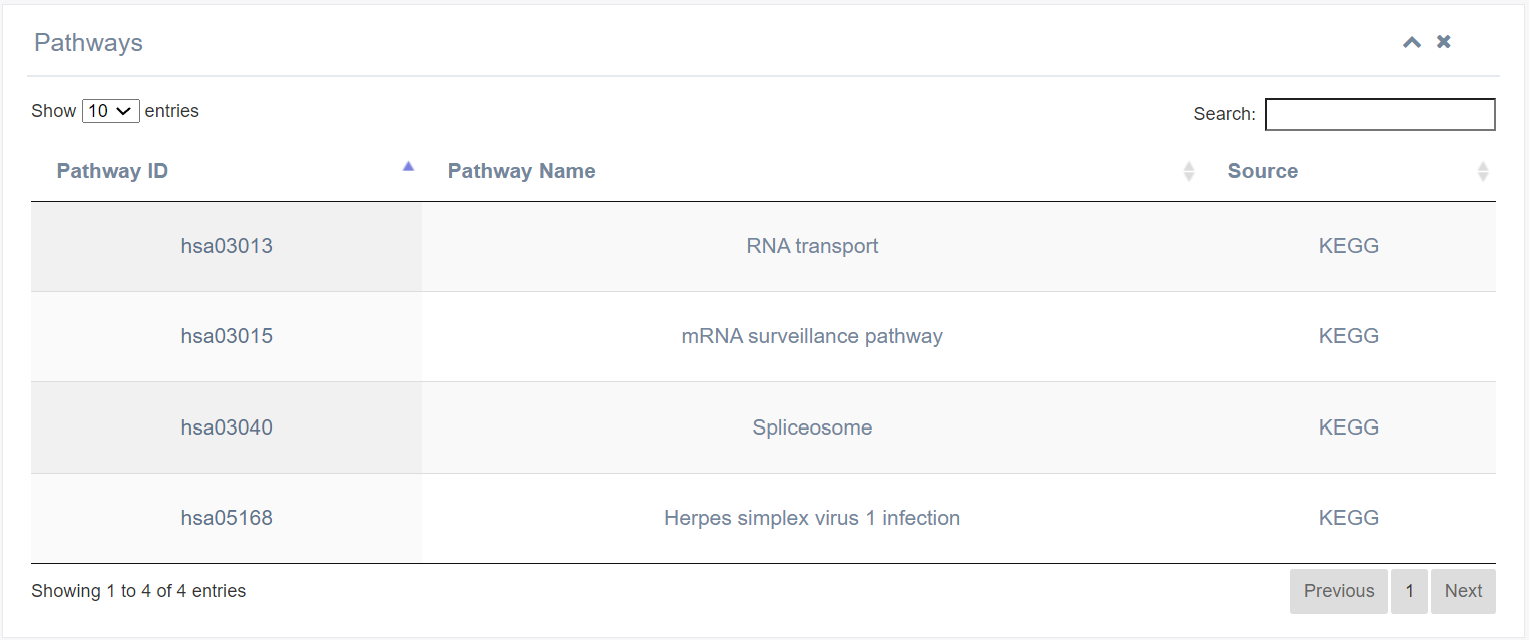
7. Pathway
This table displays signaling pathway information associated with the ALYREF protein, sourced from the KEGG database.
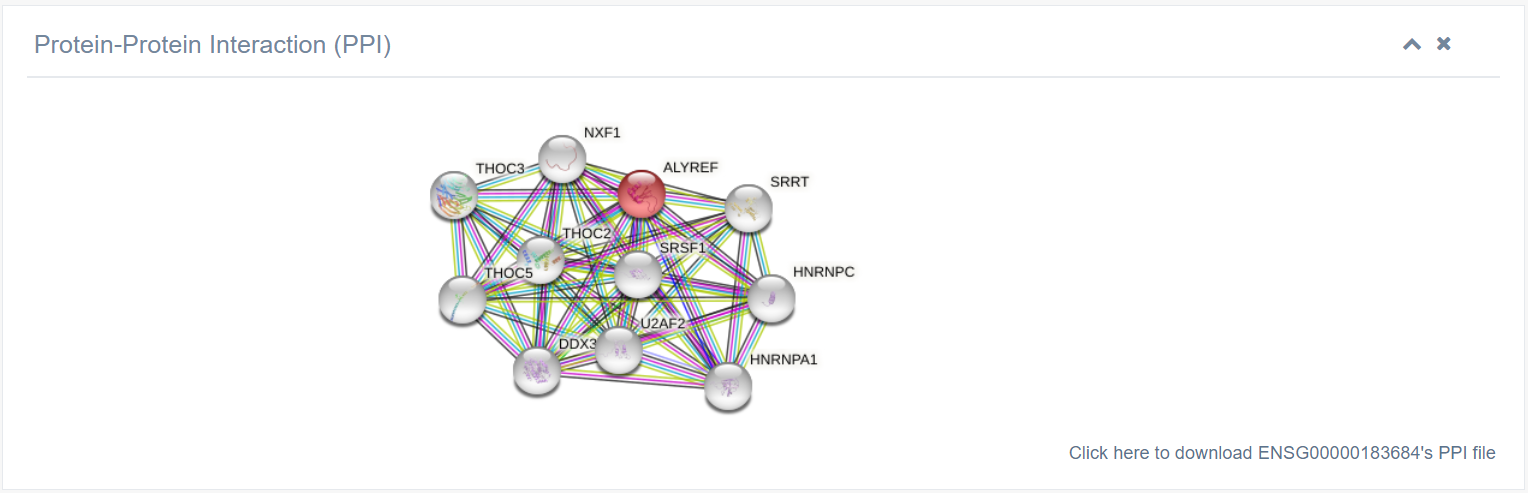
8. Protein-Protein Interaction (PPI)
Detailed interaction information can be downloaded by clicking the link in the lower right corner.
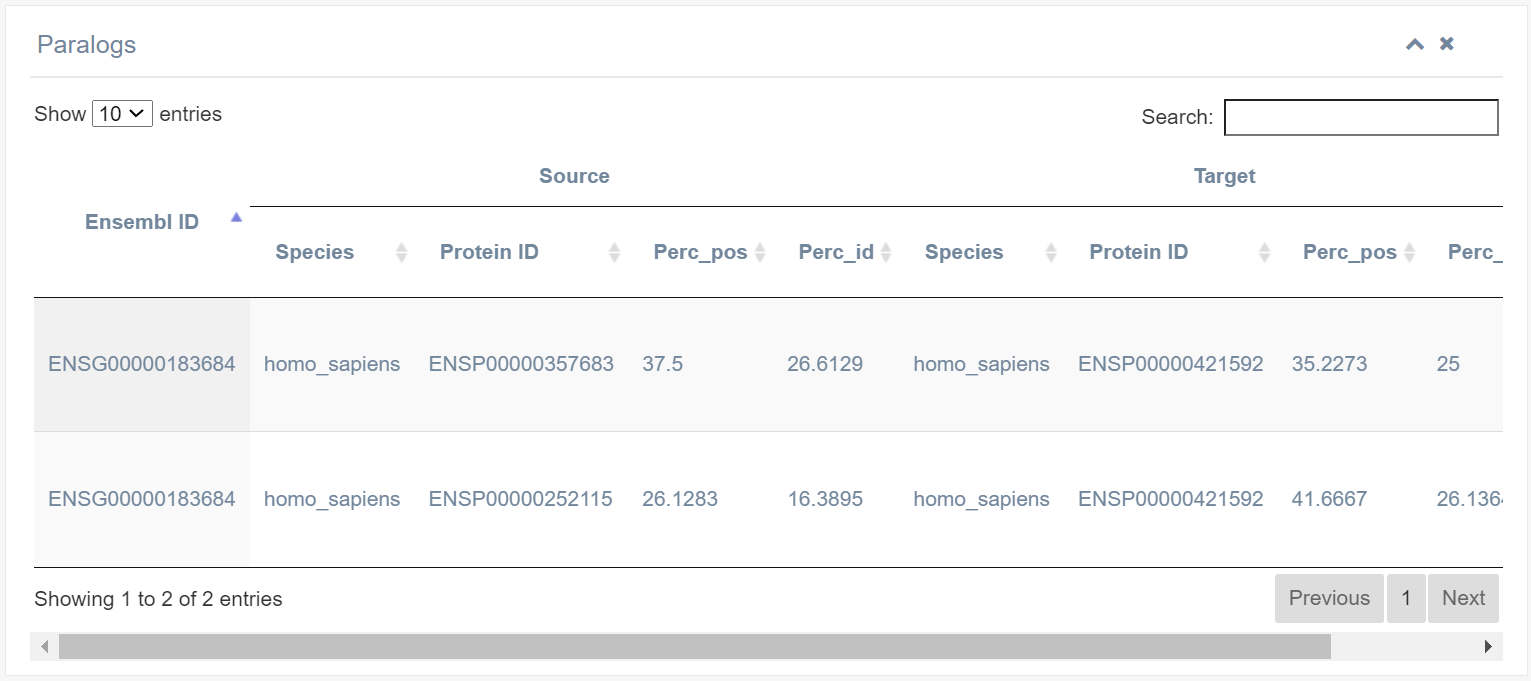
9. Paralog
This table lists all paralogs of the selected RBP. The best hit method (RBH) predicts putative orthologs of RBPs across different species using all-against-all BLASTP search with specific cutoffs (E-value ≤ 1e-6, coverage ≥ 50%, identity ≥ 30%).
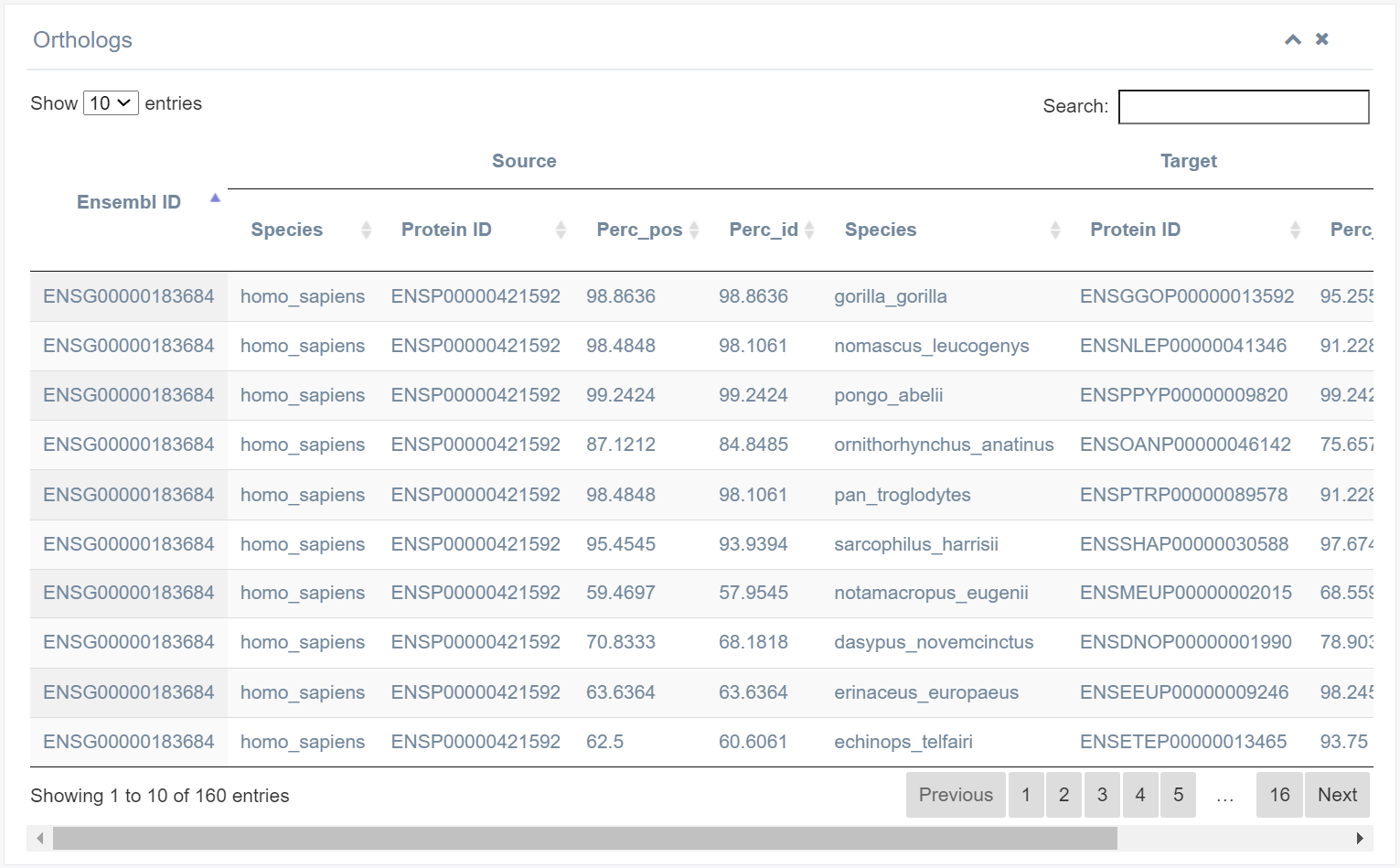
10. Ortholog
This lists all orthologs of the selected RBPs. Paralogs are predicted using the BLAST score ratio (BSR) approach. BLASTP searches are conducted in each genome using the same parameters as for searching orthologs, with a BSR cutoff set value of 0.4.
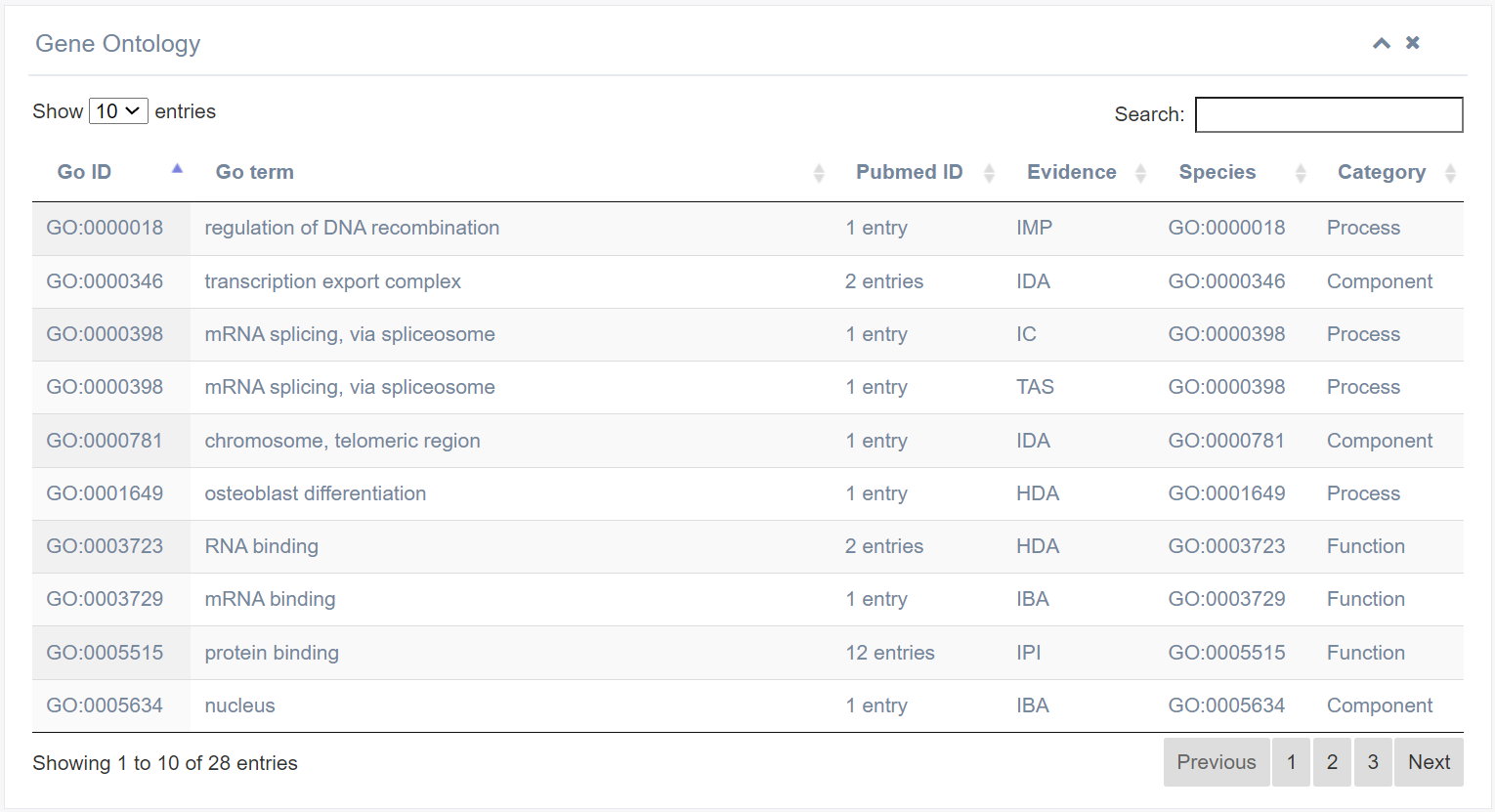
11. Gene Ontology
Annotations are parsed from the gene2go file downloaded from NCBI ftp. EuRBPDB identifies abundant RBPs from species such as human, mouse, zebrafish, yeast, fly, and worm using the hmmsearch program and manual collection in the HMMER (v3.2.1) software package.
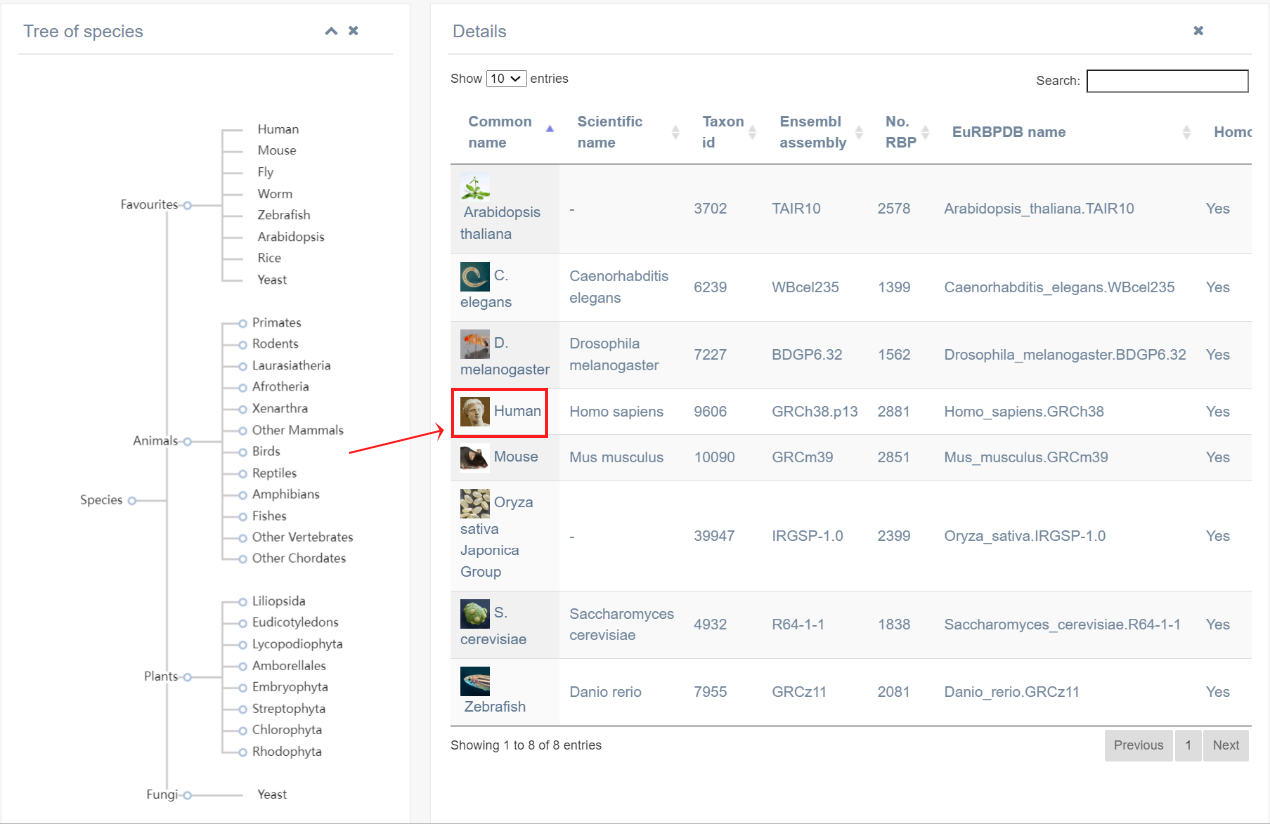
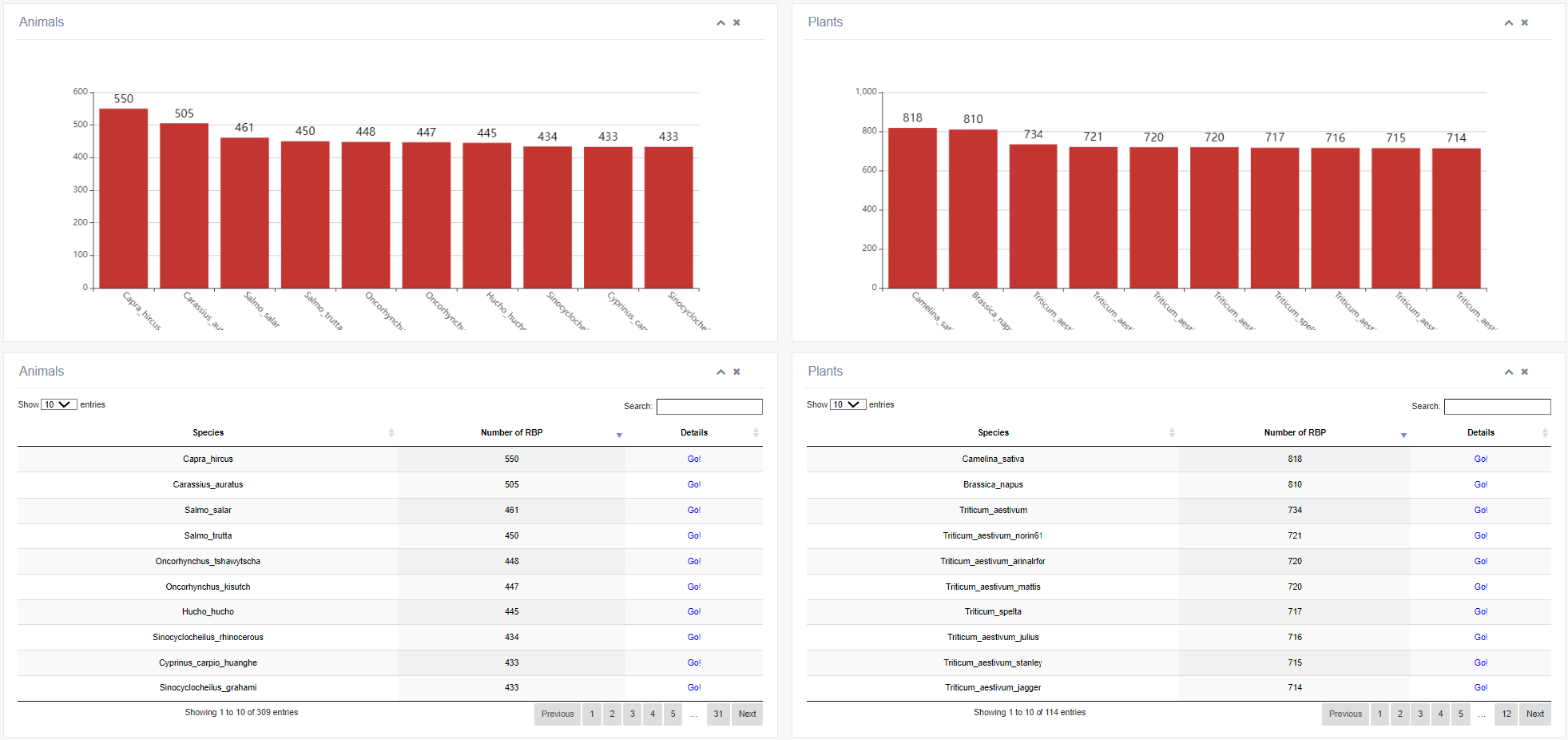
The left figure displays the species tree in EuRBPDB, while the right figure provides details for each species, including common name, scientific name, Texon ID, the number of RBPs recognized by each species, and the presence of homology.
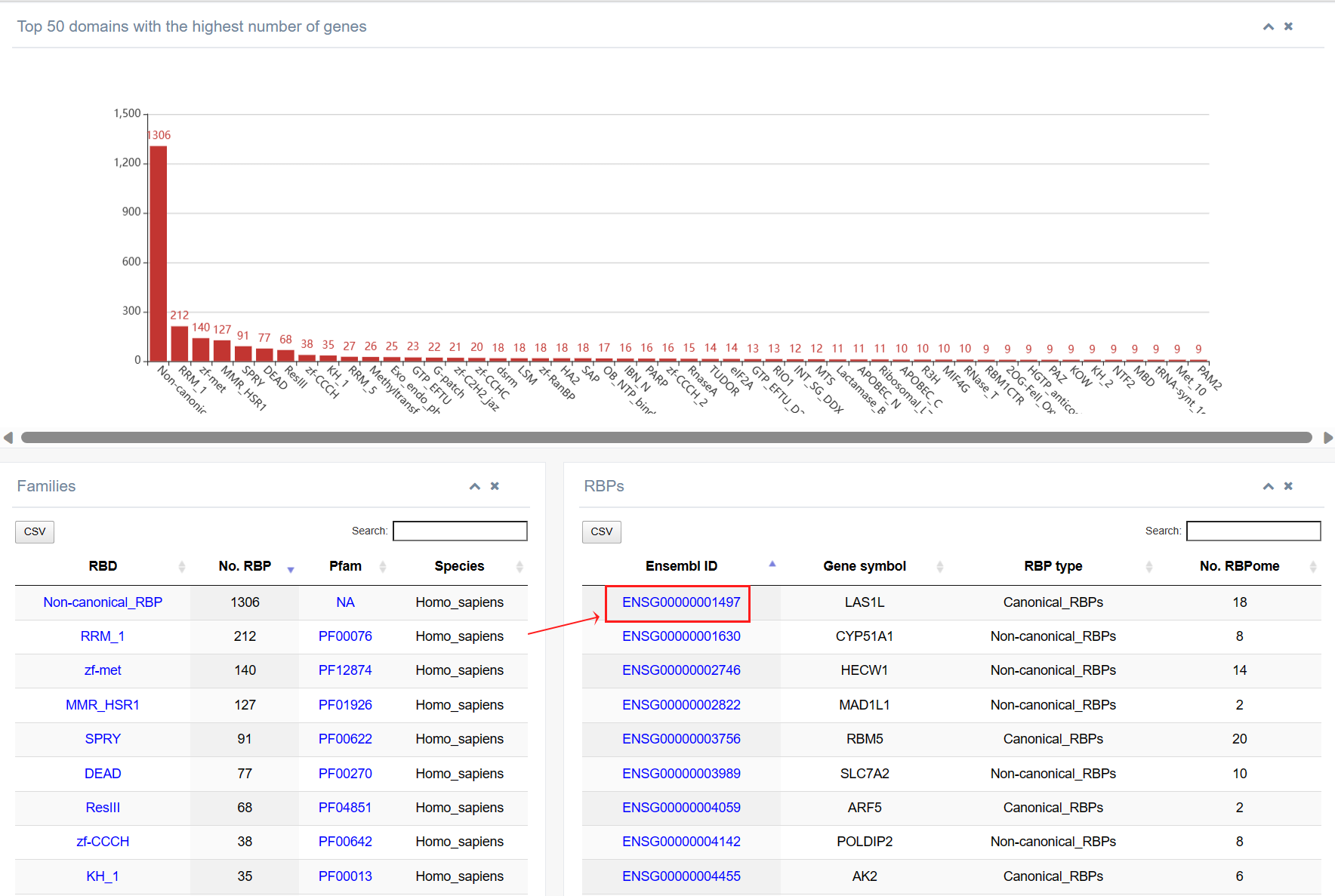
1. Taking human species for an example, by clicking on the “Common name” column, the page will return information on the top 50 RBDs with the highest number of genes. Based on the different RBD types, RBPs were divide to different RBP families. And we also provide the detailed information about these RBP families, including RBDs, number of RBPs, Pfam ID. Clicking on the “Pfam” column links to the external database - InterPro. Also, the further information on specific RBPs is also displayed, like Ensembl ID, gene symbol, RBP type and number of RBPome related studies.
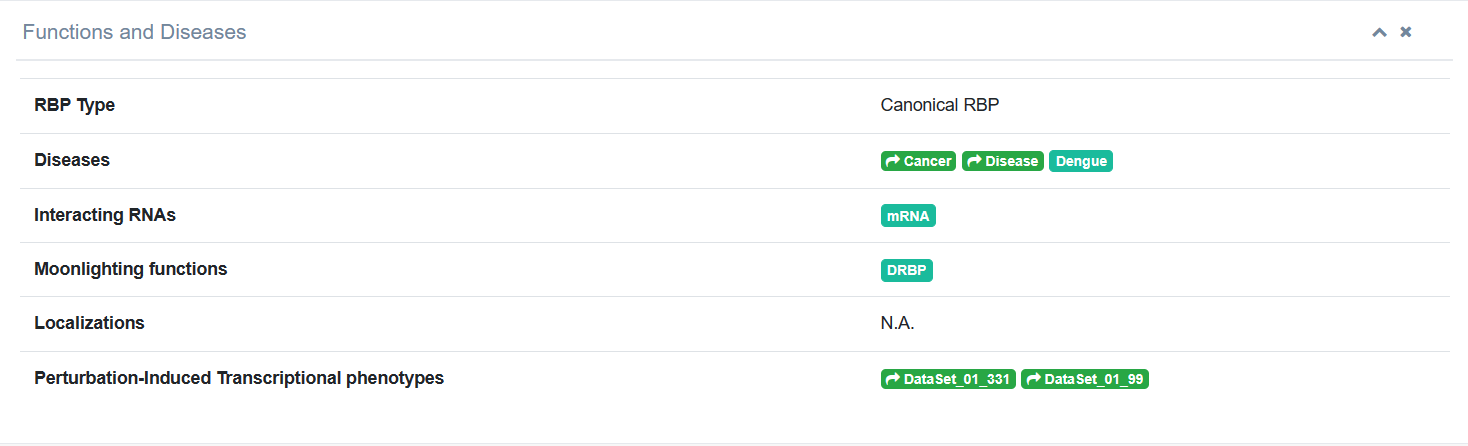
2. While clicking on the “Ensembl ID” column, page jumps to the detailed information about this RBP included in RBPWorld. We then take U2AF2 (ENSG00000063244) as an example, and users can explore the details of this gene encoding for a RBP: Functions and diseases This section shows an overview of the various SUBJECTS of RBPs included in RBPWorld, including RBP type (whether it contains known RBDs), linkage to disease (including Cancer/Genetic diseases/Viral Infections, etc.), interacting RNA types, Multifunctional RBPs (whether it has moonlighting functions, such as DRBP/E3/DUB/Transmembrane protein/Kinese), Condensate RBPs and information on Perturbation-Induced Transcriptional phenotypes. Click on the specific hyperlink to jump to the detail page.
RBPs are characterized and classified based on sequence-specific RBDs. If an RBP contains only one type of RBD, the RBP family is named after the RBD domain. If an RBP contains multiple types of RBDs, it is placed in the family with the smallest E-value in the RBD prediction. Non-canonical RBPs are grouped into the non-canonical RBP family. EuRBPDB collects data on 663 RBP families, and the figure above shows the top 50 RBP families with the largest number of genes.
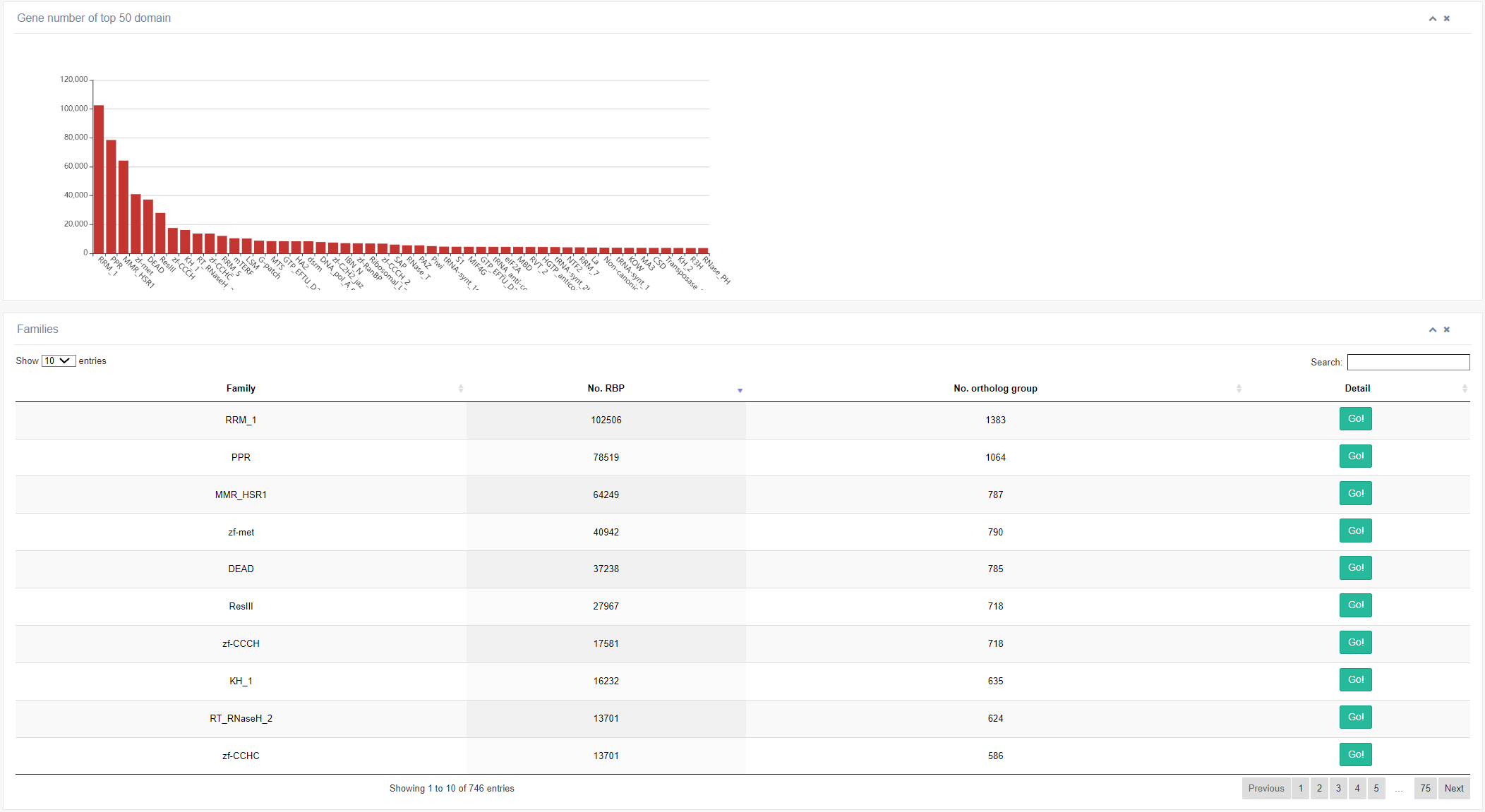
Clicking the 'Go!' button in the table below queries information about the of a specific RBP family across different species (animal or plant).
This module includes four sub-modules: Cancers, Genetic diseases, RNA virus diseases, and Drugs. Users can conveniently explore the disease associations of RBPs using this module.
I. Cancers
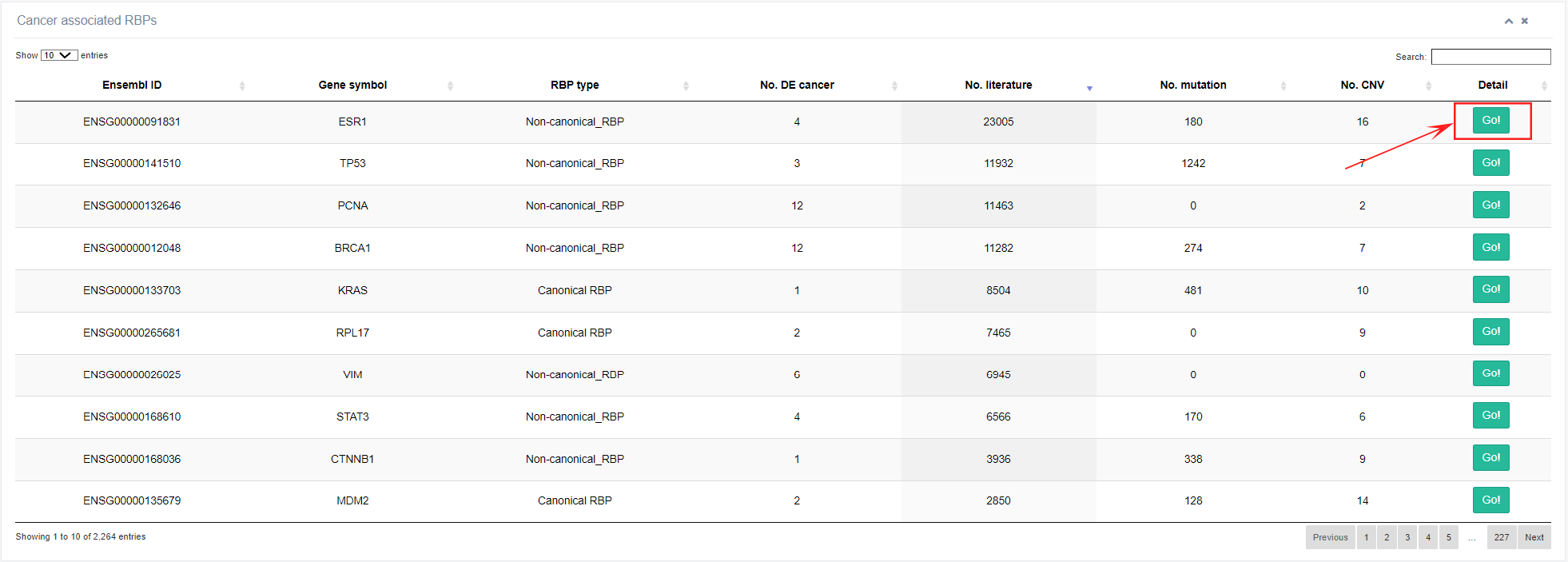
This page provides information on cancer-associated RBPs, including RBP name, type, The number of cancer type in which RBP differentially expresses, number of published publications, number of mutations, and number of CNVs. Clicking on the last column will lead to the details of a specific RBP.
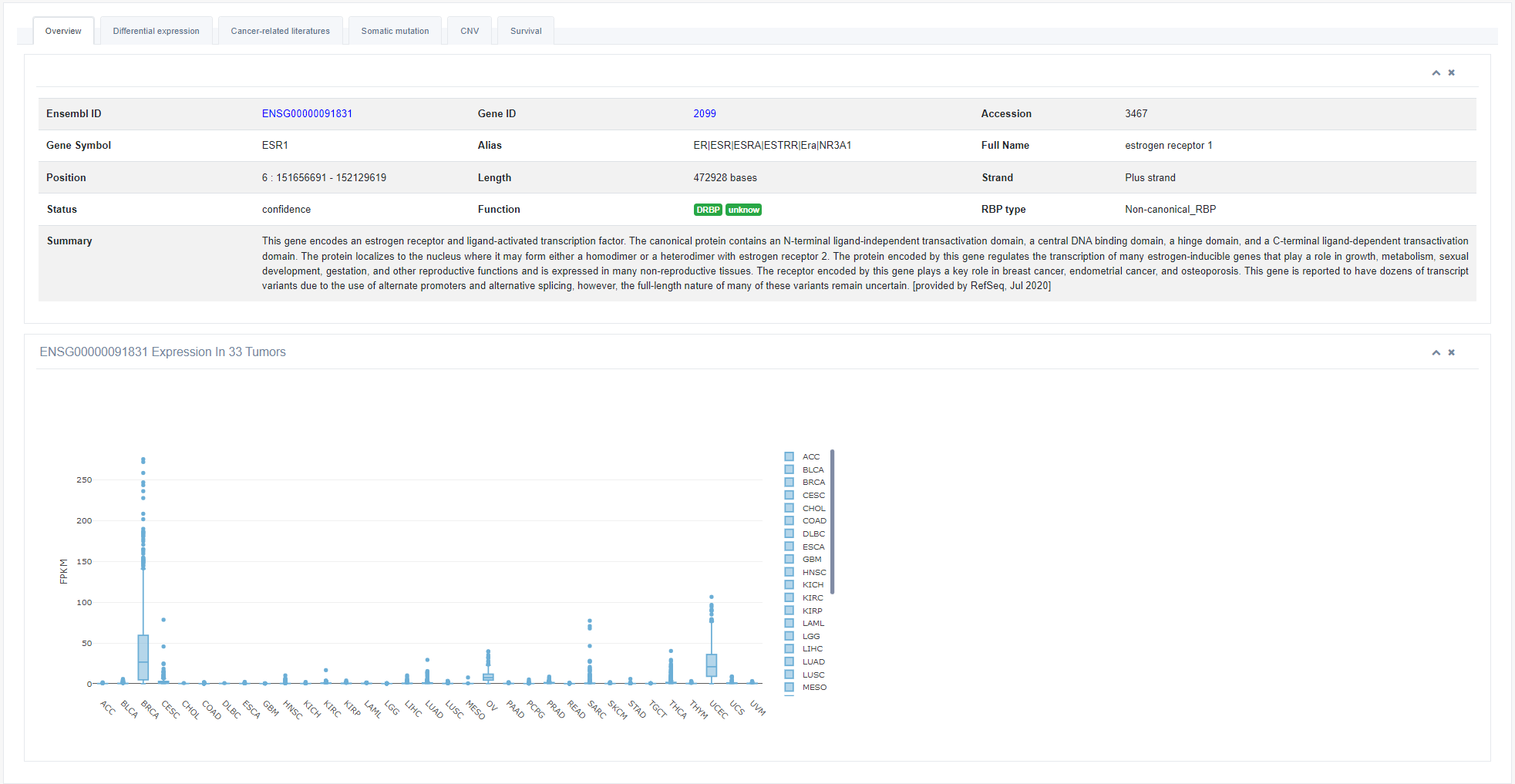
1. Overview
As a start, this page shows the basic information of RBP and the expression level of RBP in 33 cancers. Users can add or remove samples from the boxplot by clicking the sample name on the right panel.
2. Differential Expression
The boxplot shows the expression level of an RBP in tumor and normal tissues. Only cancers exhibiting differential expression of selected RBP are shown in boxplot.
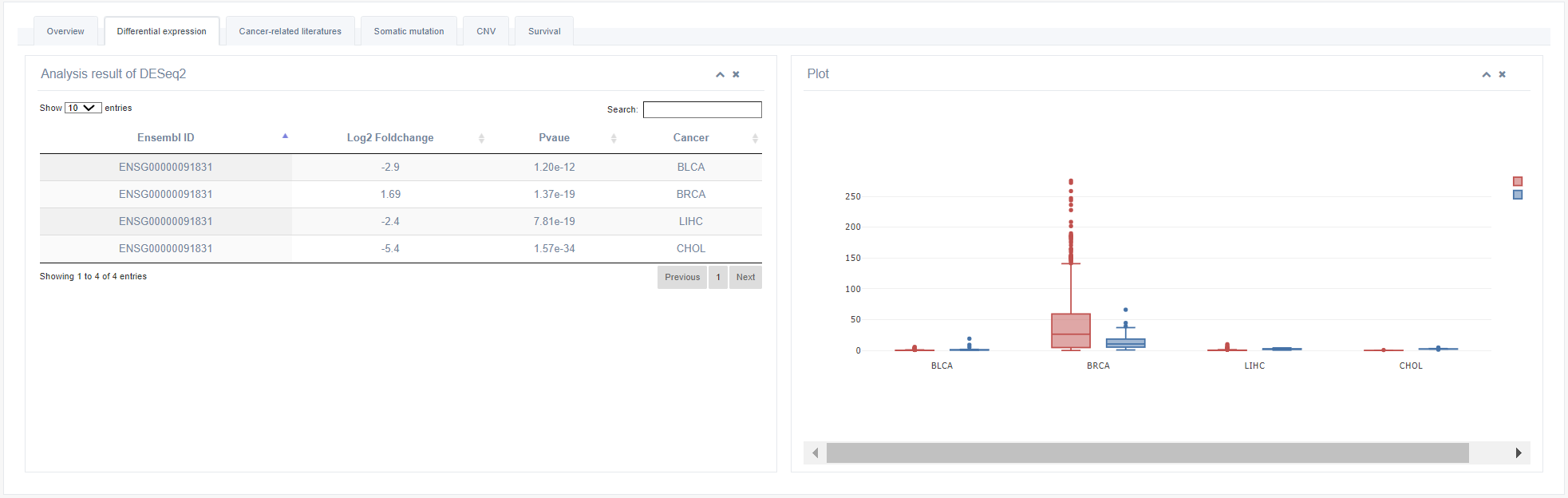
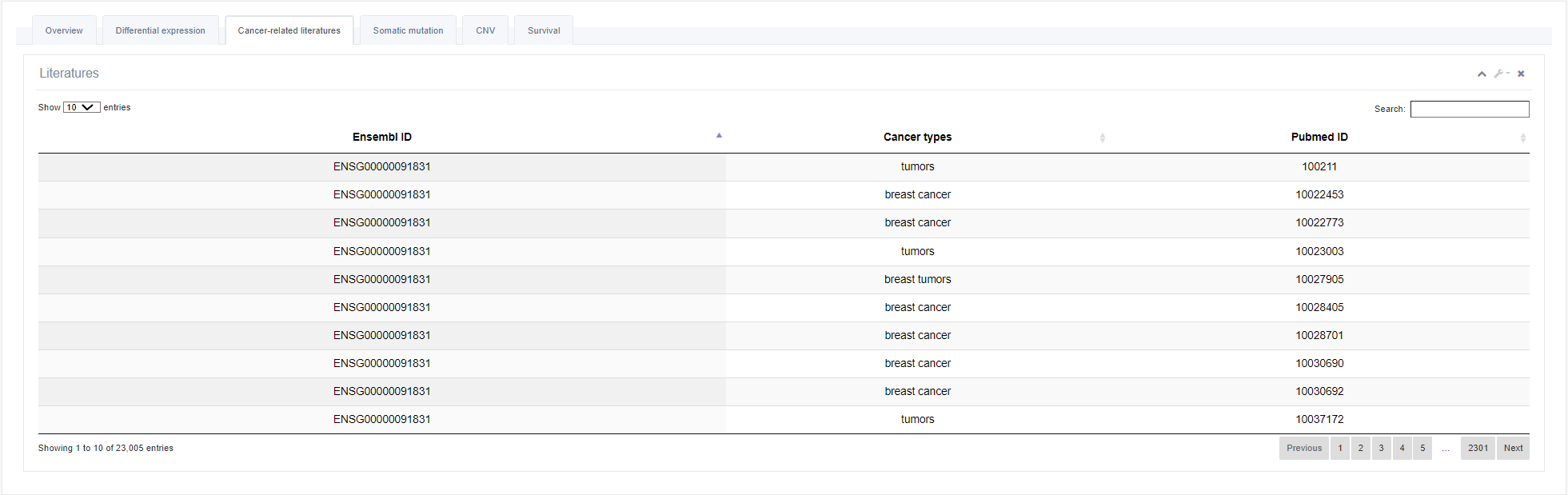
3. Cancer-related literatures
All information currently reported in the literature on this RBP is presented on this page.
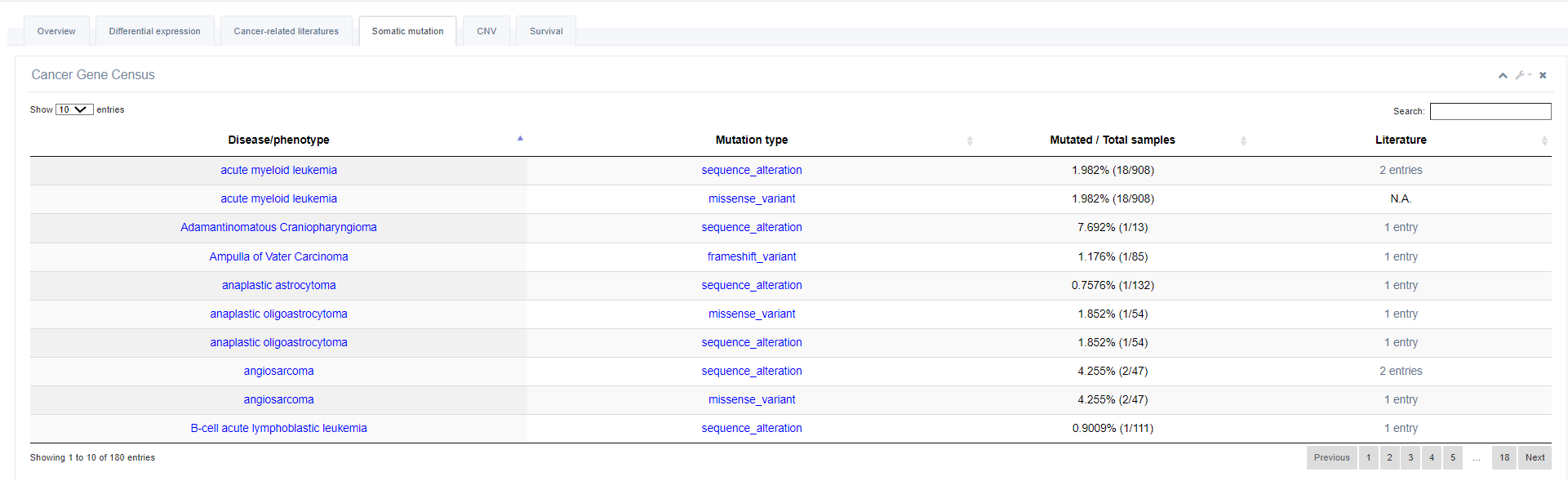
4. Somatic mutations
The table lists all mutation an RBP has. Users can obtain the information of mutation type, disease/phenotype, sample and literature in each cancer from the table.
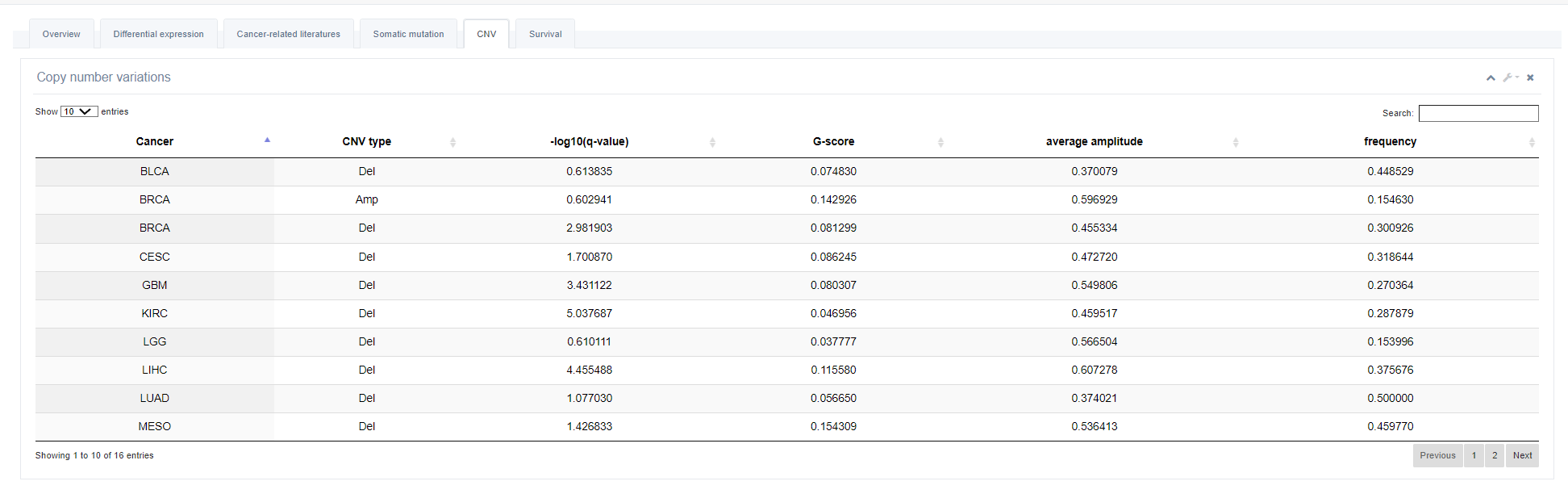
5. Copy Number Variation (CNV)
This part lists all cancers with deletion or amplification of selected RBP.
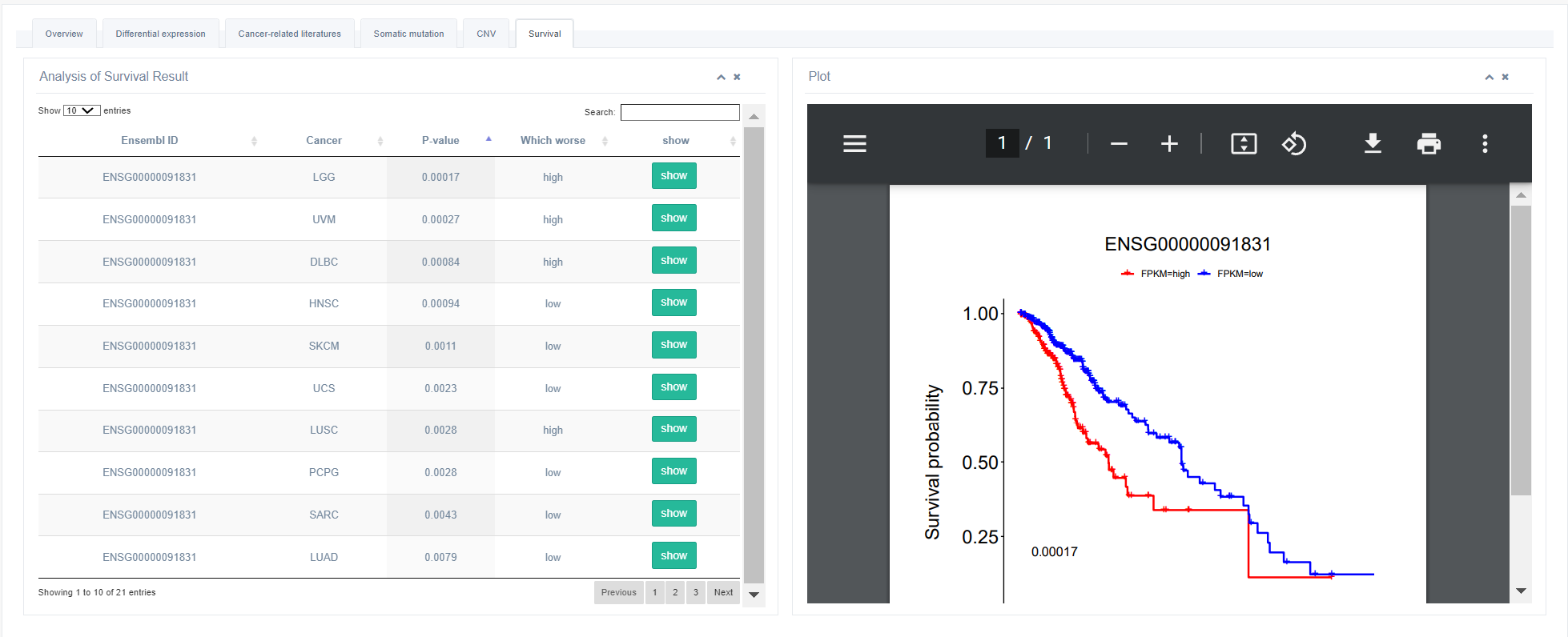
6. Survival Analysis
This part lists all cancers with significantly different survival state between high expression group and low expression group of selected RBP. Clicking the "Show Figure" will generate a Kaplan-Meier survival plot which can be downloaded as PDF format.
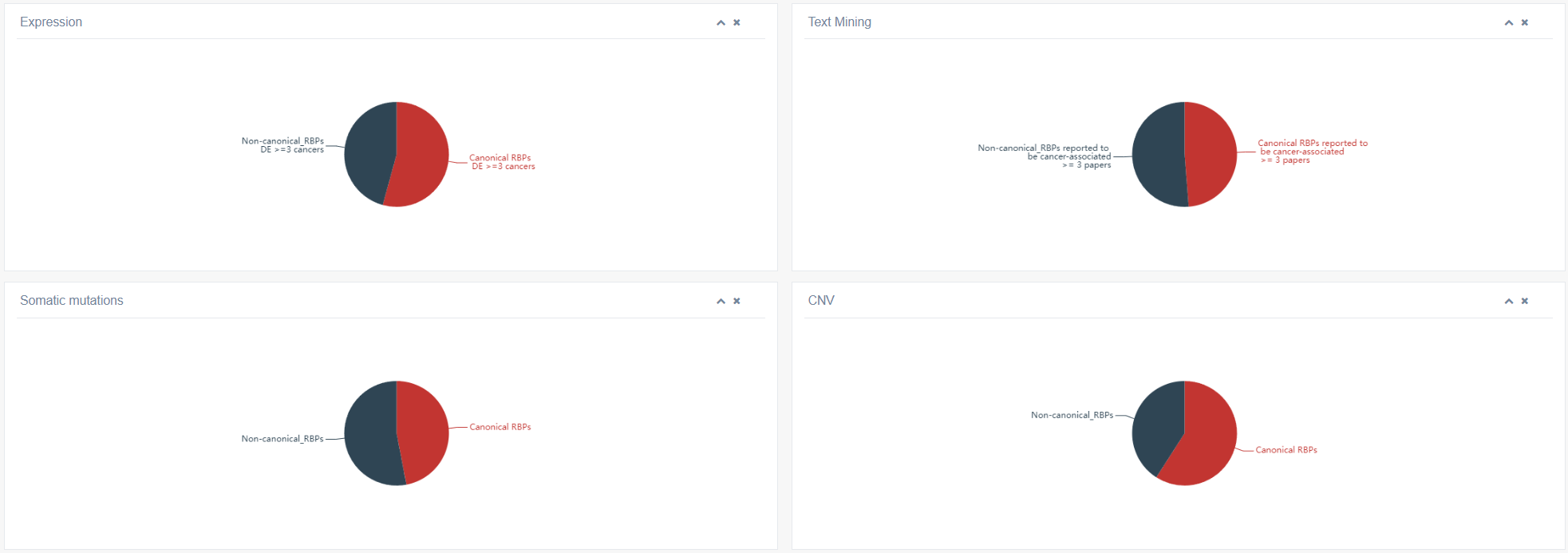
The figure below illustrates the distribution of non-classical and classical RBPs under various conditions, such as differential expression in ≥3 tumors, reported in ≥3 publications, somatic mutations and CNVs status.
II. Genetic diseases sub-module
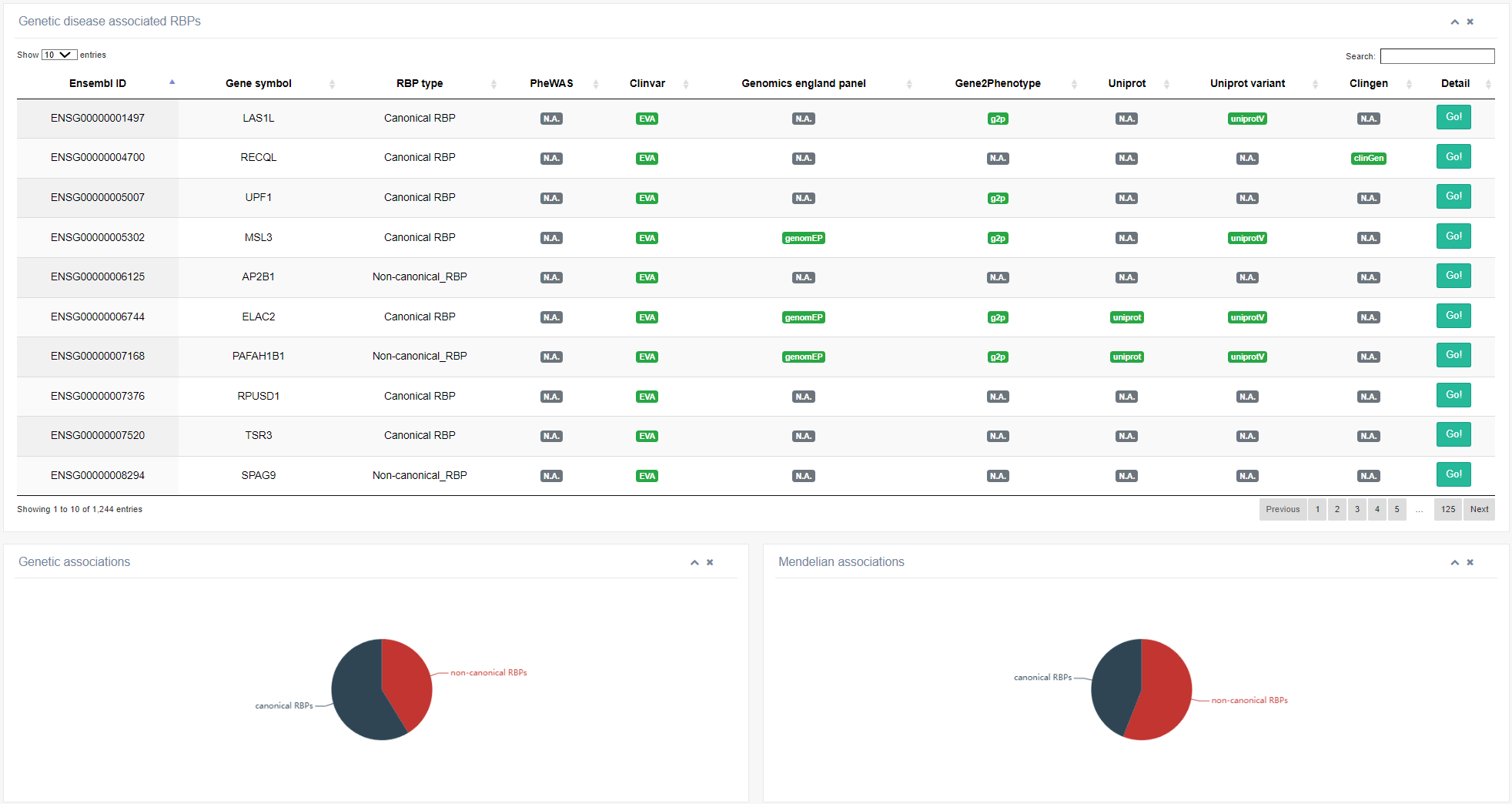
RBPs play a role in various human genetic disorders. Through analysis of disease-associated data from public databases such as GWAS, ClinVar, ClinGen, Cosmic, etc., 1342 RBPs have been found to be associated with human genetic disorders. This page provides the complete list of genetically associated RBPs, and users can explore genetic evidence for RBPs in the "Details" table. Clicking the "Go!" button for each RBP leads to detailed information on each evidence.
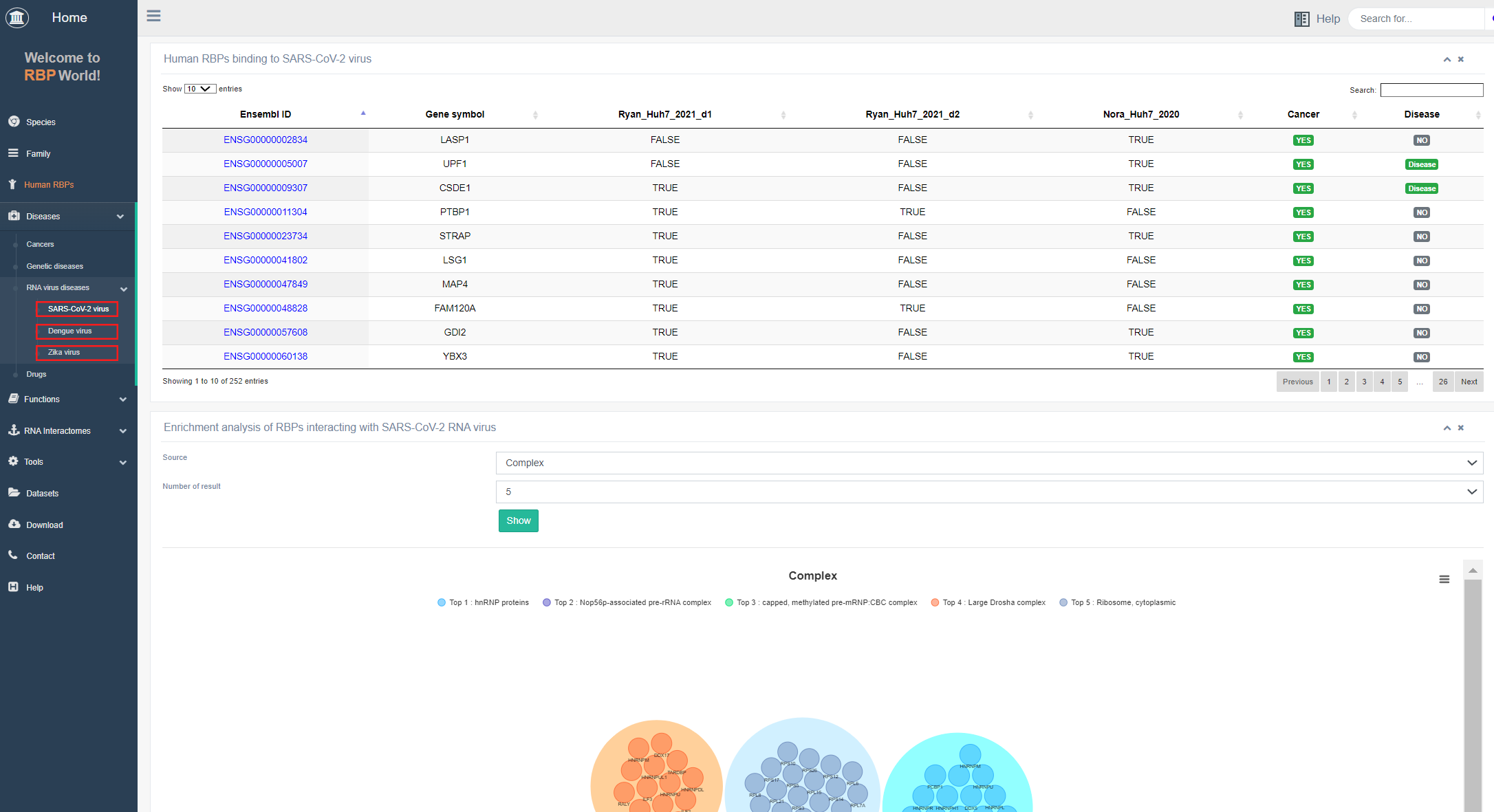
III. RNA virus sub-module
EuRBPDBV2 includes 252 RBPs associated with SARS-CoV-2, 591 with Dengue, and 206 with Zika RNA viruses, identified through probe-based RNA techniques. These RNA virus-associated RBPs can assist in identifying relevant host proteins functioning as RBPs and contribute to understanding the complex interactions involving these RBPs and the respective viruses. In the RNA virus submodule, users can find a comprehensive list of RBPs binding to SARS-CoV-2, Dengue, and Zika RNA viruses, enabling exploration of the derived complexes.
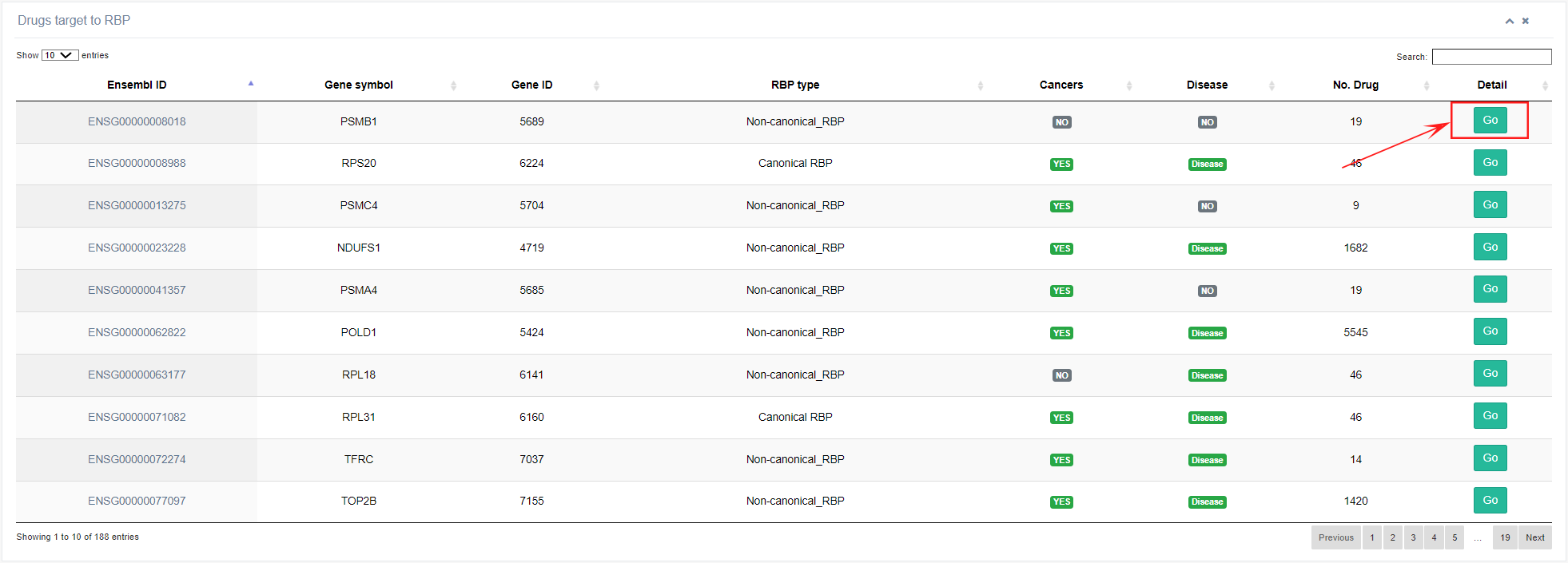
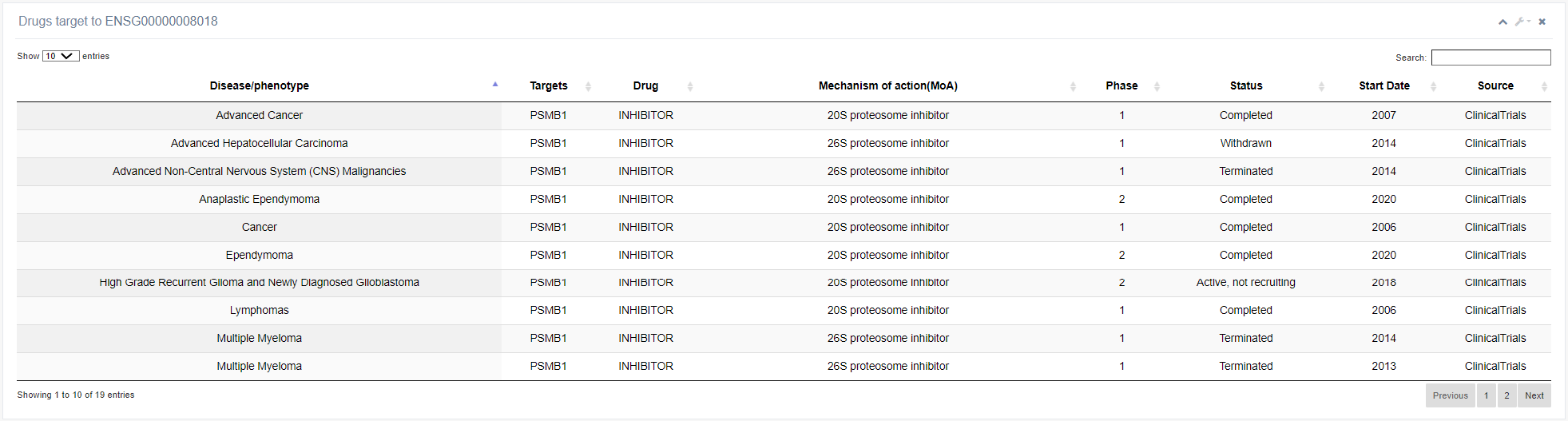
IV. Drug
In this section, users can access targeted drug information for selected RBPs. By clicking the "Go!" button, the website will provide information on the type of disease, drug name, mechanism of action, project status, and initiation time of clinical trials conducted for the selected RBP.
This module facilitates exploration of the functions of RBPs. Increasing evidence suggests that the roles of RBPs extend beyond their canonical RNA-binding activities. EuRBPDBV2 has revealed 336 RNA-binding metabolic enzymes, 343 DNA-binding transcription factors, and 100 E3 ubiquitin ligases within its database. The Function module allows users to investigate the RNA targets of various RBPs.
1. Perturbation-Induced Transcriptional Phenotypes (PITP)
This module describes phenotypic changes after perturbation of the genes encoding RBPs. Users can narrow down by selecting "Family" or "RBP" on the left, or directly enter the name of the RBP they would like to find in the search box on the right to explore.
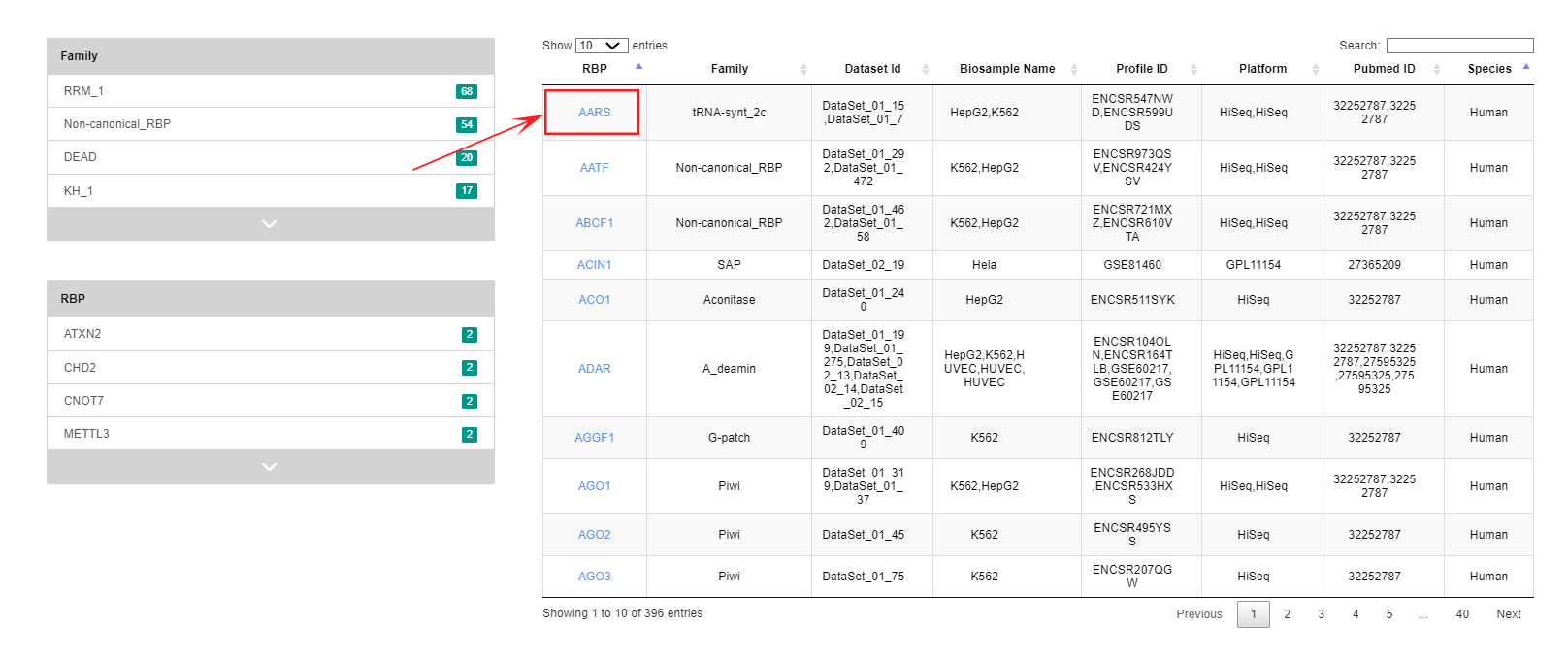
Clicking on the RBP name in the leftmost column jumps to the details page, which begins with an overview of the RBP's basic information; Taking the " AARS" protein as an example, users can explore the PITP details of this RBP.

The table describes all the target genes of the RBP, containing information such as gene symbol, the number of datasets in which RBP differentially expresses.
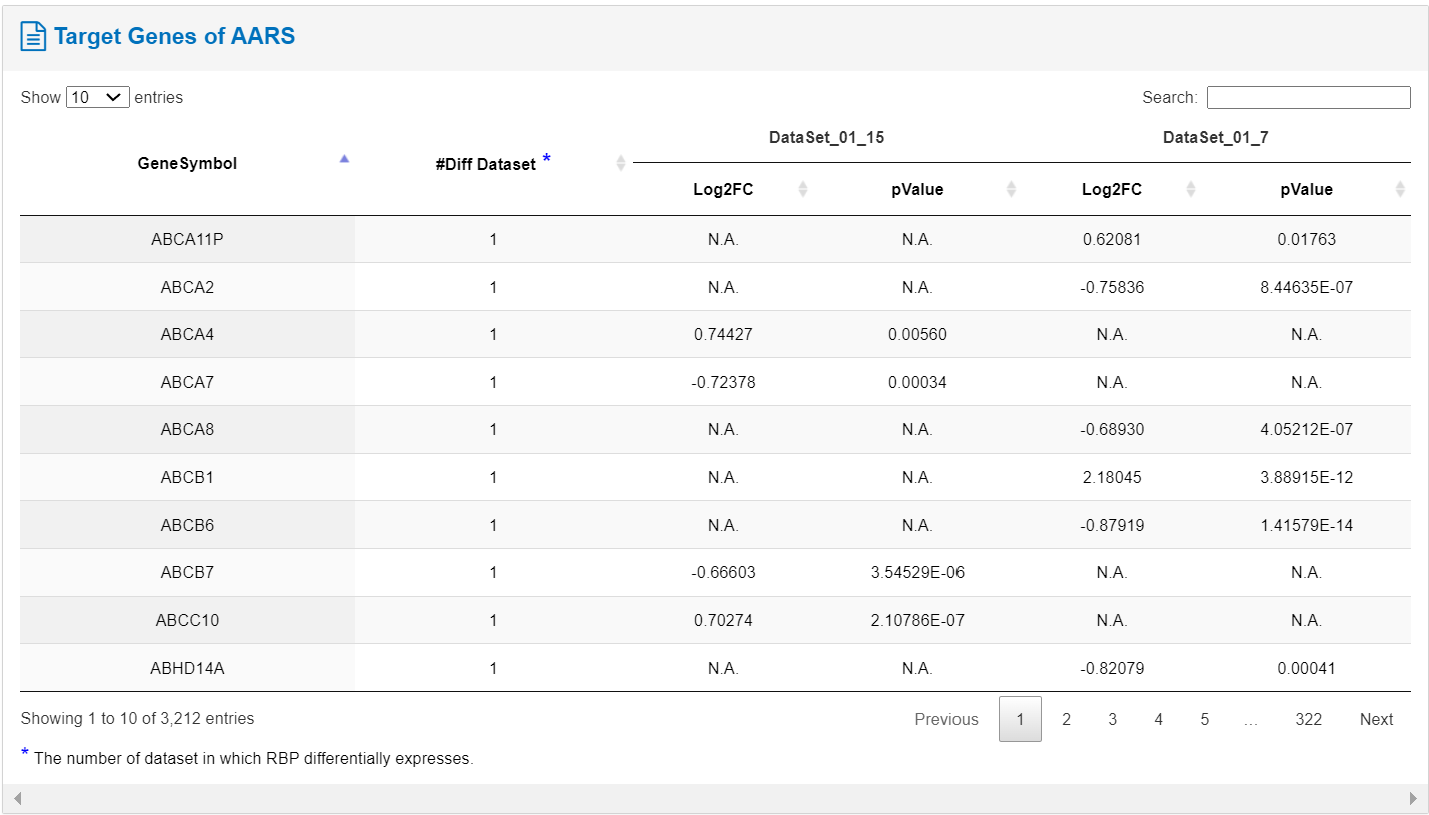
The accompanying figure illustrates the gene bound by the RBP, indicating whether target gene expression is up- or down-regulated following RBP perturbation. These data have been obtained through CLIP data analysis.
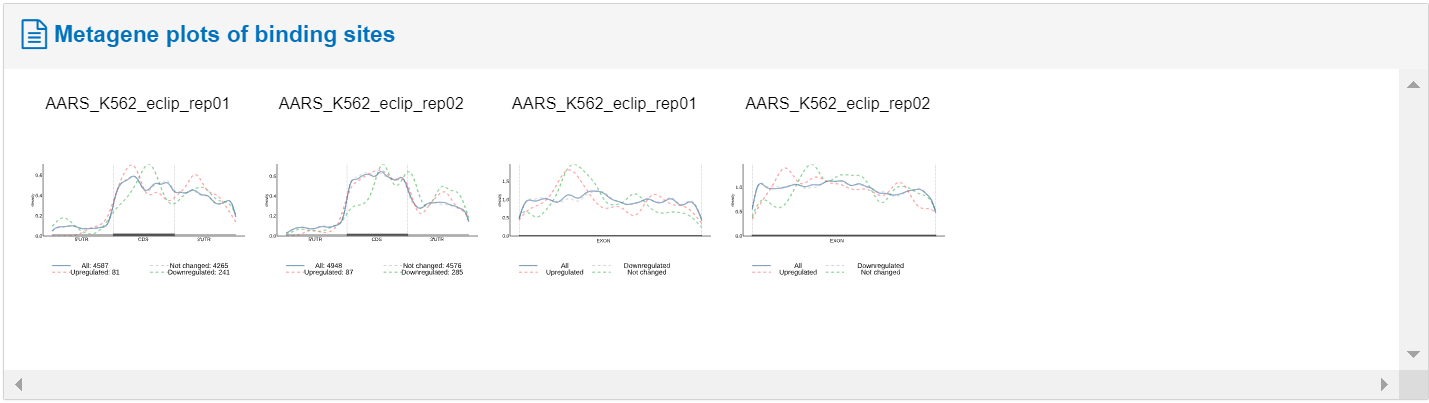
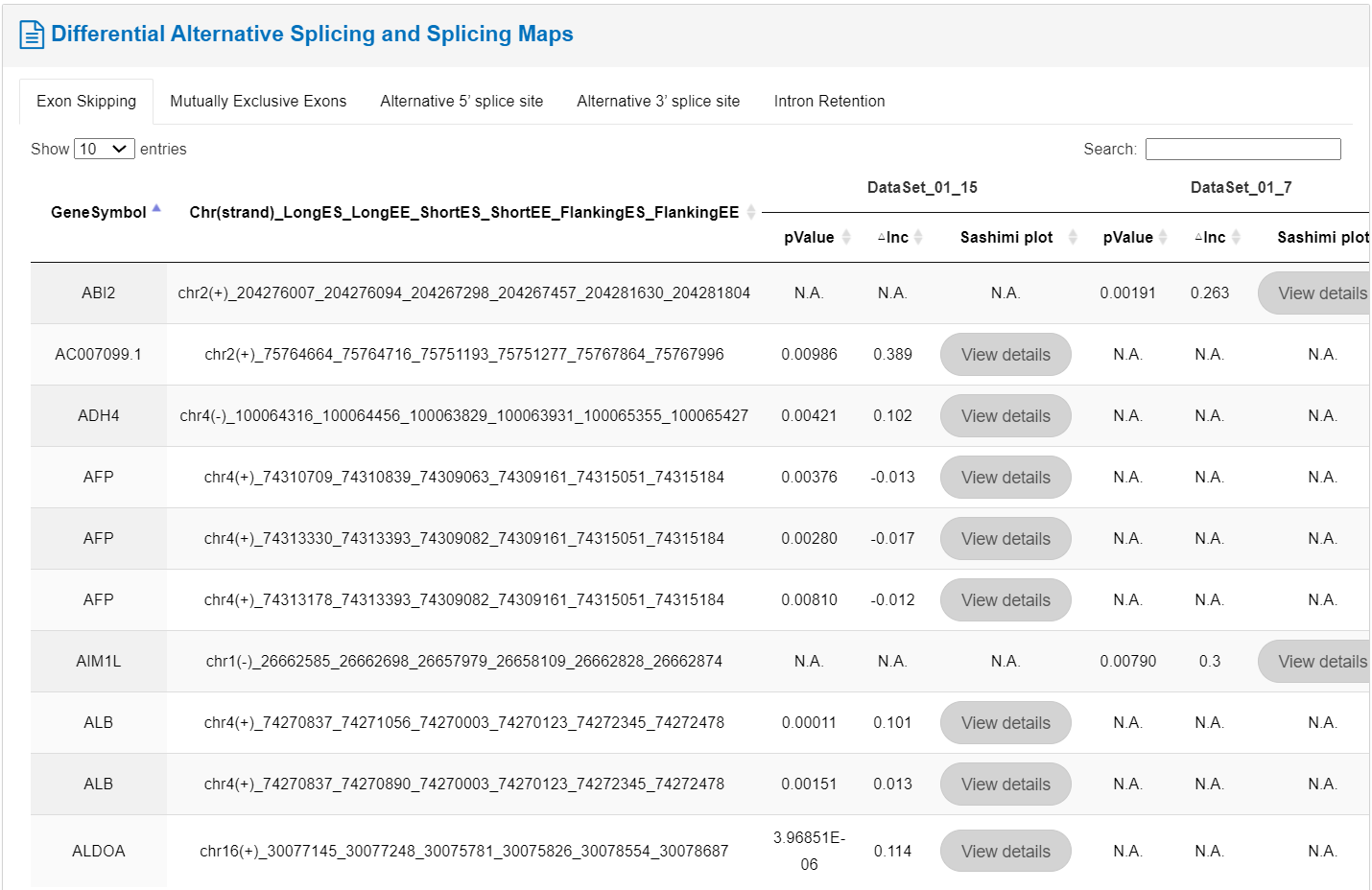
Additionally, the table presents differential variable splicing and splicing profiles of RBP target genes, encompassing phenomena such as exon skipping, mutually exclusive exons, alternative 5' splice sites,' splice sites, and intron retention. Clicking the rightmost viewing of high-resolution images.
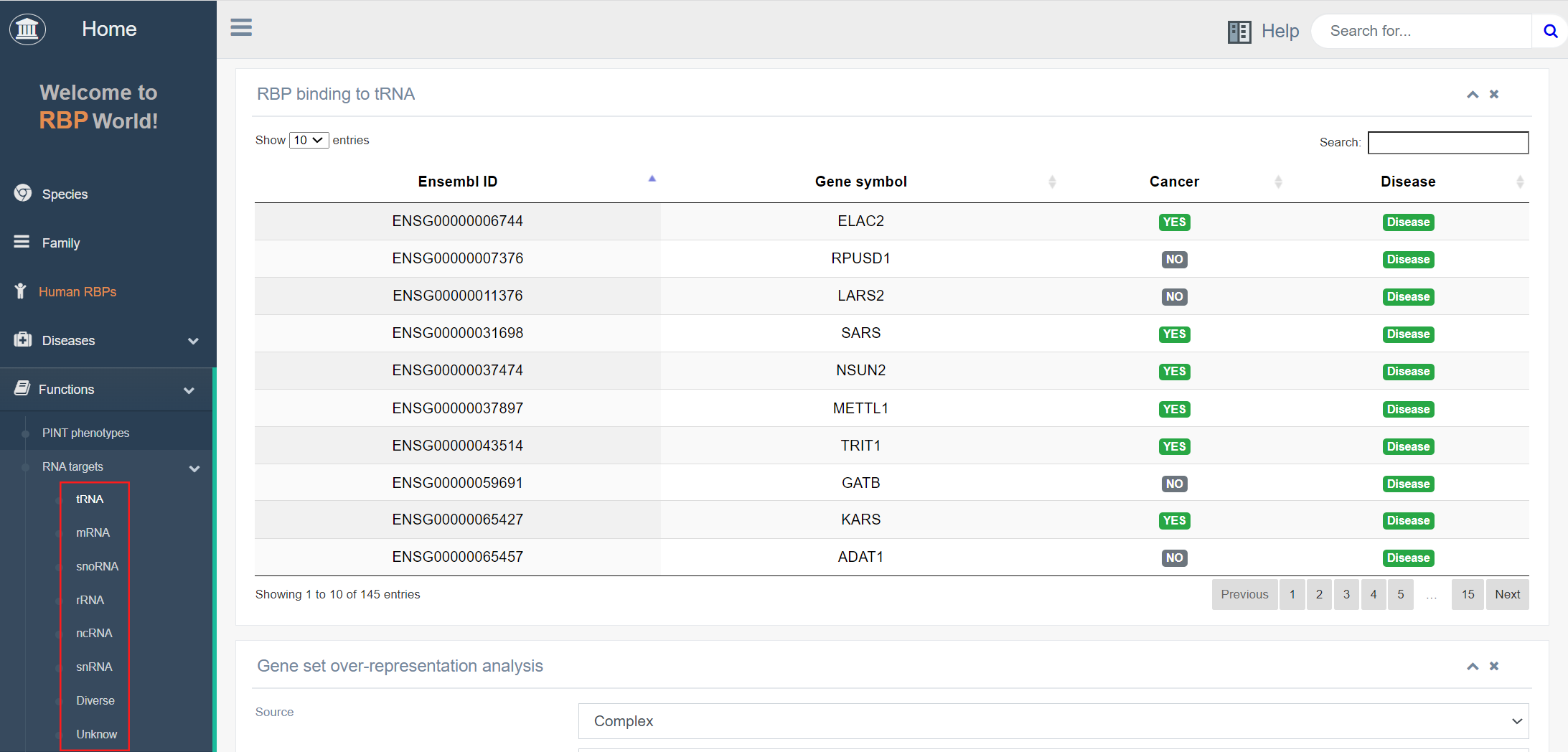
2. RNA Targets
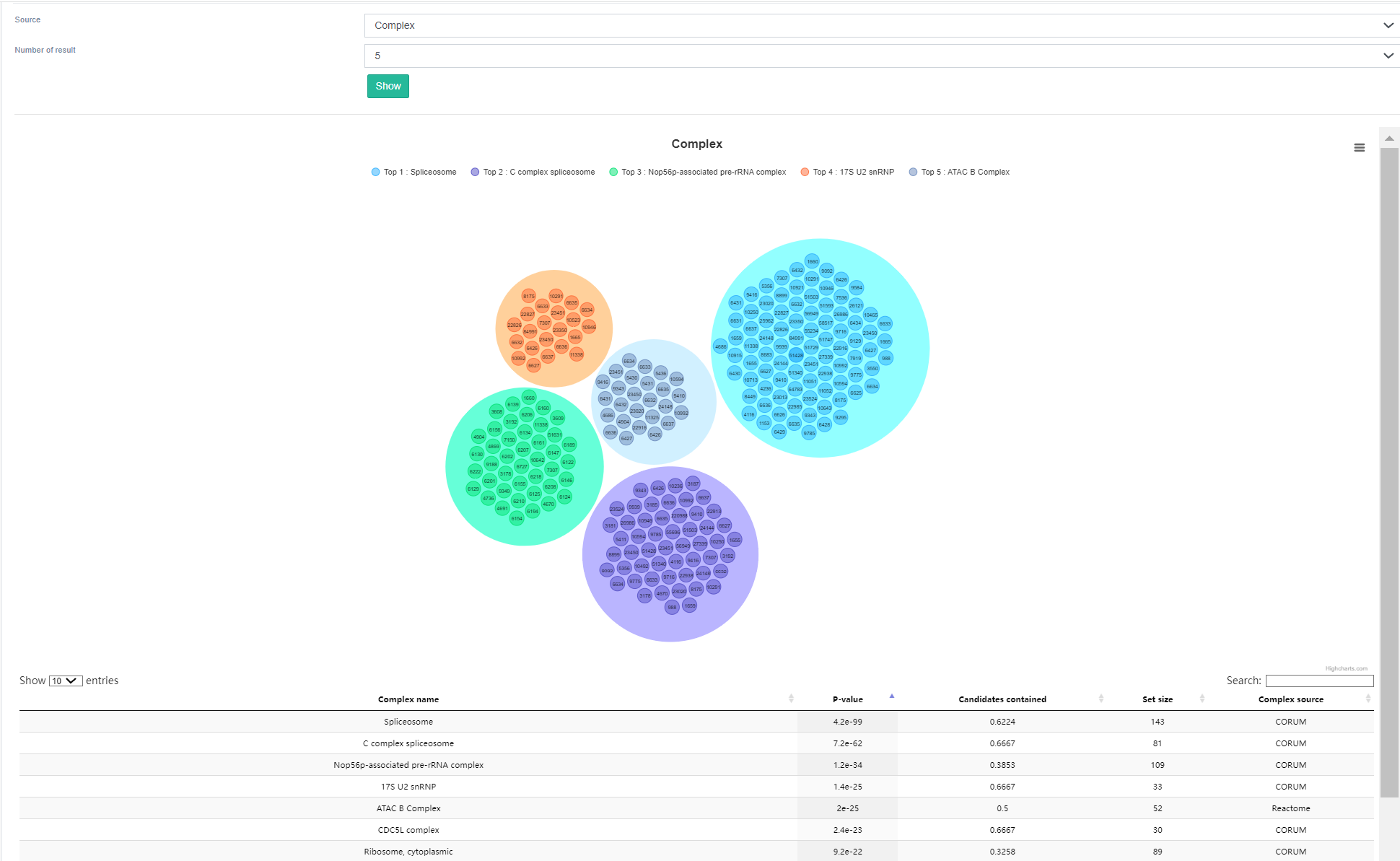
This module describes the binding capacity of RBPs to various types of RNAs, including tRNA, mRNA, snoRNA, rRNA, ncRNA, snRNA, or other unknown RNAs. For instance, focusing on RBPs that bind to tRNA, the top image on this page displays the collection of 145 RBPs identified by EuRBPDB. It also indicates whether these RBPs are associated with cancer or other diseases.
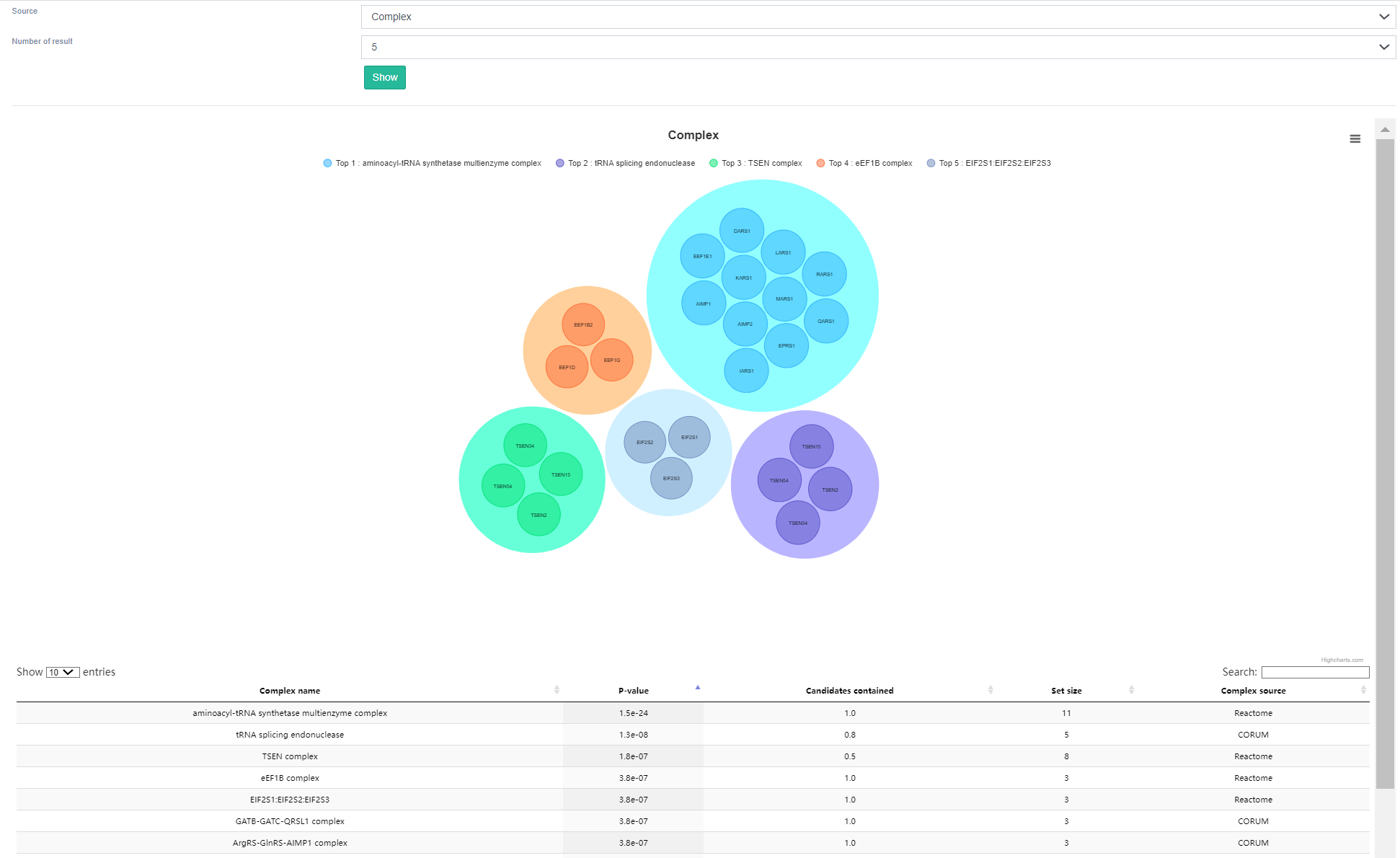
Through ORA enrichment analysis of the functions and pathways, the lower panel exhibits the top 5 scoring complexes as results of gene set over-representation analysis. Interestingly, it is evident that RBPs binding to tRNA are largely enriched in the hnRNP protein complex. The function of hnRNP proteins primarily involves mRNA metabolism, regulation of RNA transcription, splicing, and translocation processes, which closely align with the role of tRNA in promoting gene expression.
The RBPs section of this module, which binds to other specific types of RNAs, functions in a similar way to "tRNA";
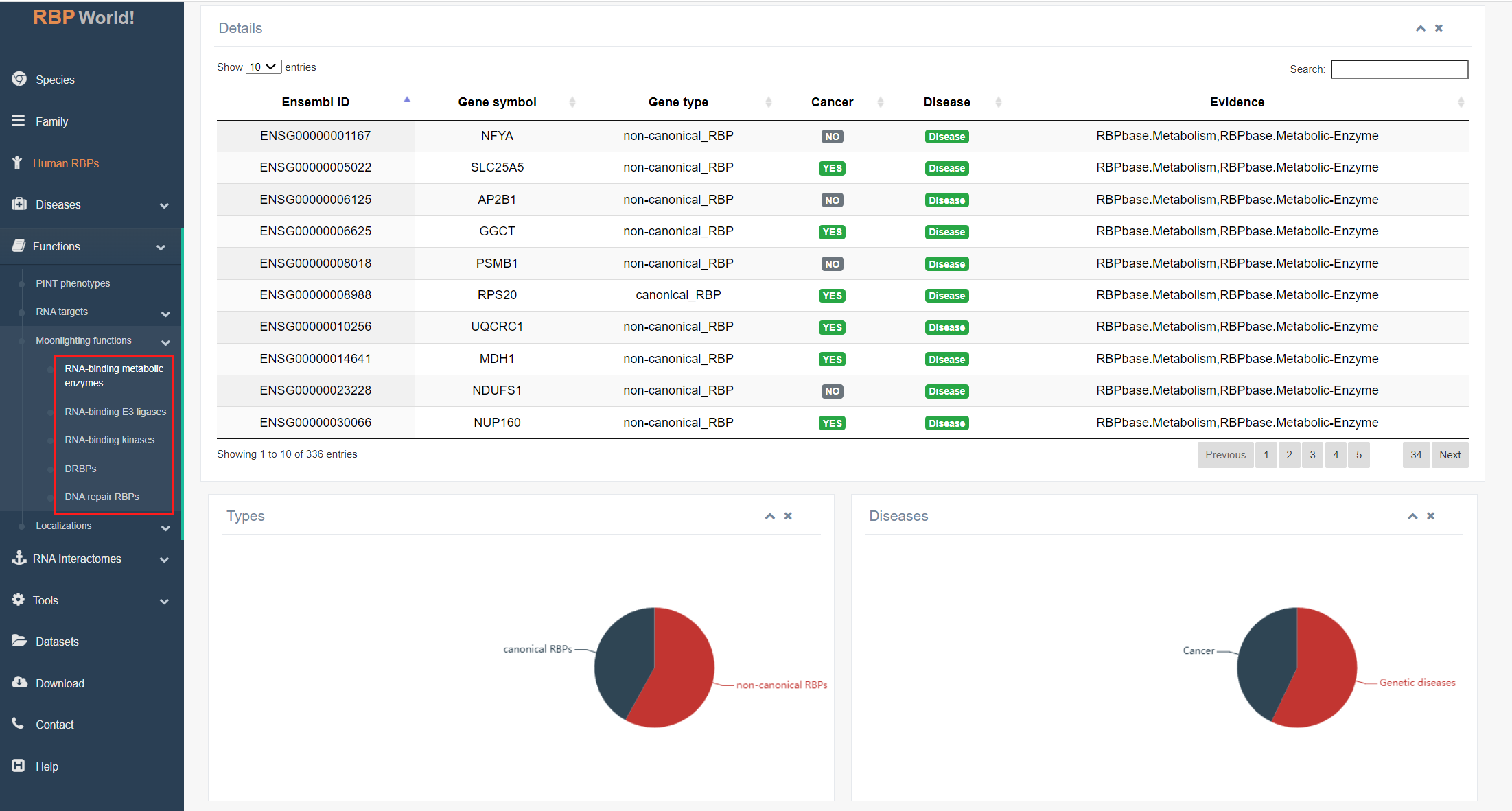
3. Moonlighting Functions
This module highlights RBPs with specific roles, encompassing 336 RNA-binding metabolic enzymes, 100 RNA-binding E3 ligases, 45 RNA-binding kinases, 343 double-strand RNA binding proteins (DRBPs), and 56 DNA repair RBPs. Users can explore whether their RBPs of interest are involved in these particular processes.
The upper table on this page provides comprehensive details about the RBPs, including gene symbols, gene types, cancer or disease associations, and supporting evidence. The bottom graph the percentages of non-classical/classical RBPs and disease-related/non-disease-related RBPs among all RBPs.
4. Localizations
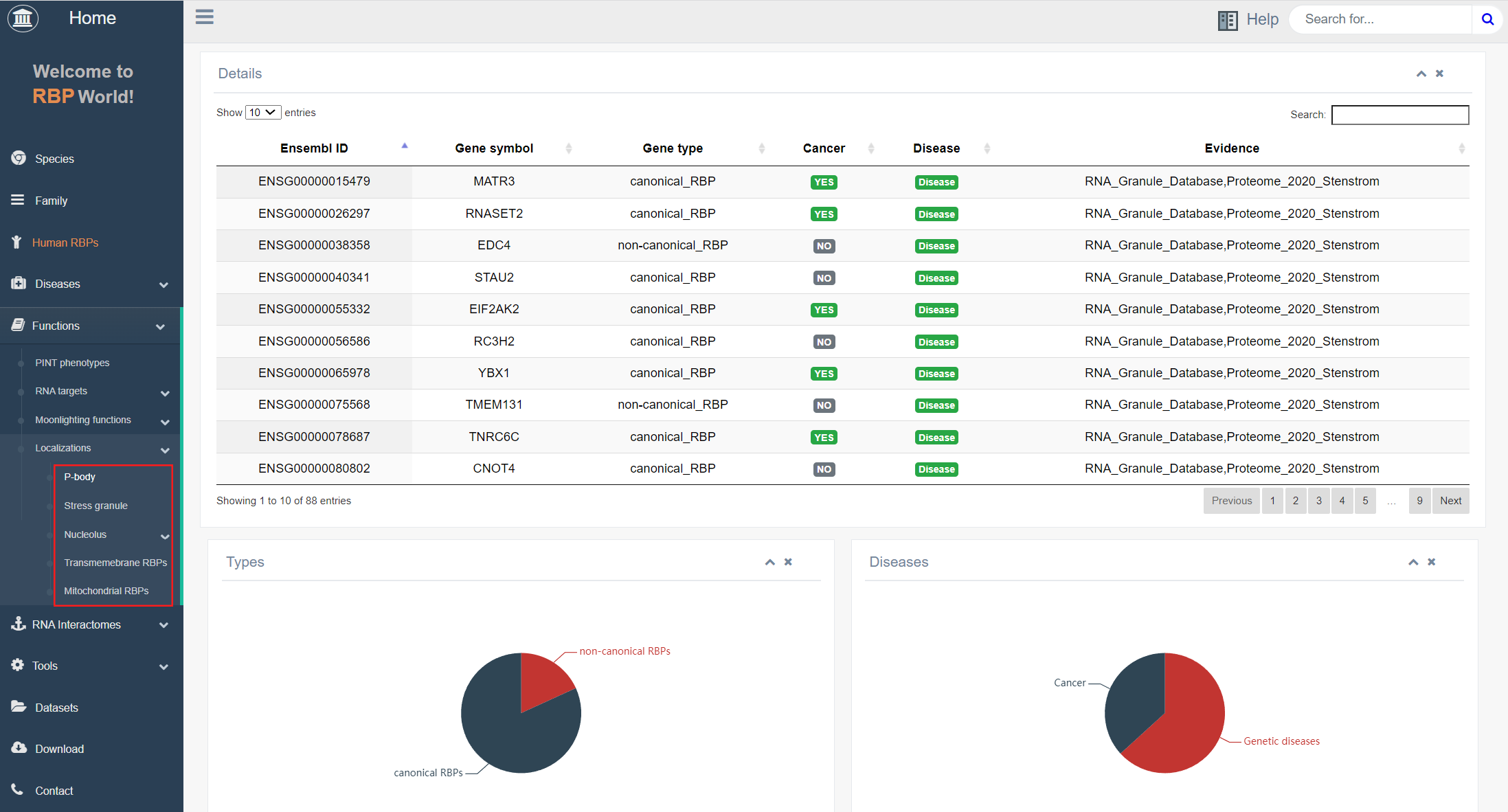
In this module, users can discover RBPs involved in the assembly of specific cellular structures, such as P-bodies, stress granules, nucleoli, transmembrane RBPs, and mitochondrial RBPs, that possess specific intracellular localizations. Users can determine whether their RBPs of interest are associated with these related structures.
The top table on this page presents detailed information about the RBPs, including gene symbols, gene types, cancer or disease associations, and supporting evidence. The lower graph illustrates the proportions of non-classical/classical RBPs and disease-associated/non-disease-associated RBPs among all RBPs.
5. RNA Interactomes
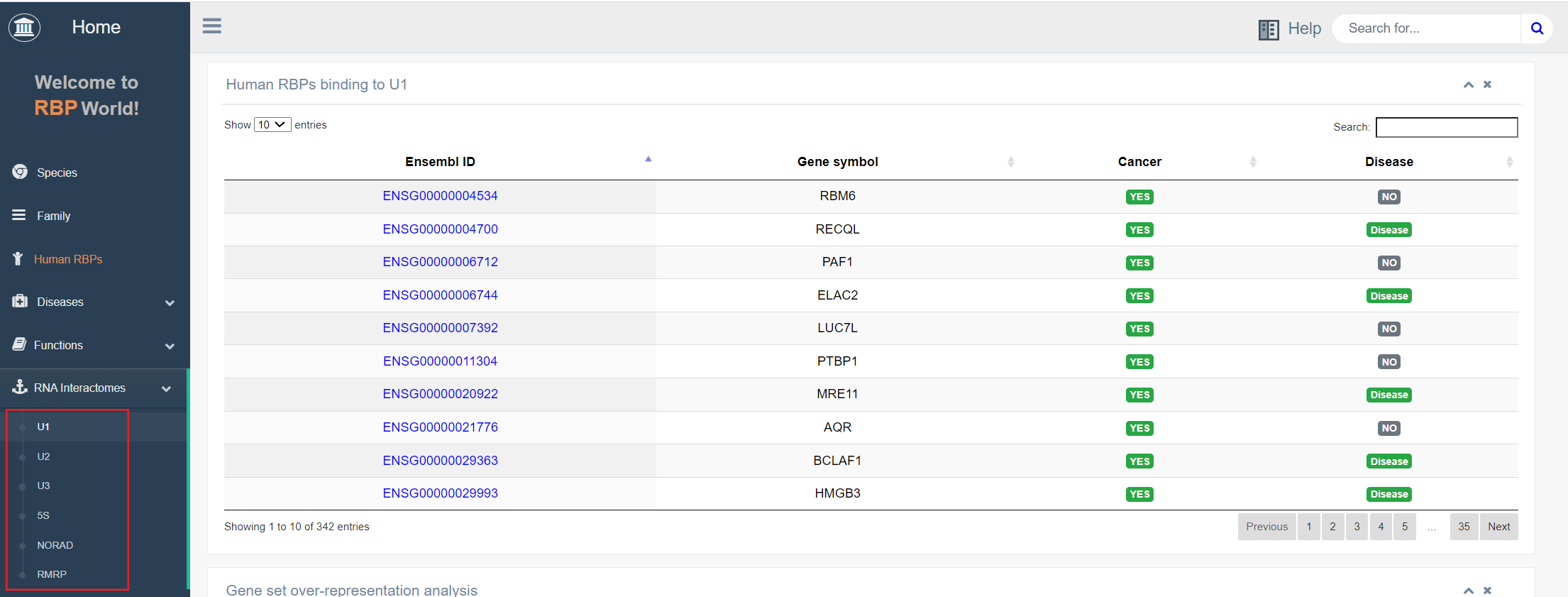
RBPs play essential roles in cellular activities by binding to specific RNAs, facilitating RNA function execution. This section describes RNA interactions involving specific RNAs, such as snRNA U1, U2, and U3, ribosomal 5S RNA, non-RNA activated by DNA damage (NORAD), and RNA-processing endoribonuclease (RMRP).
Taking RBPs that bind to U1 RNA as an example, the top panel on this page shows all 342 RBPs that bind to U1 RNA collected by EuRBPDB, and also describes whether they are cancer-associated RBPs or disease-associated RBPs.
After conducting an ORA enrichment analysis of the functions and pathways involving these RBPs, the Gene Set Over-representation Analysis results in the figure below effectively highlight the top 5 scoring complexes. Notably, RBPs bound to U1 RNA are primarily enriched in the spliceosome pathway, which bears relevance to snRNA function.