Welcome to
RBP World!
Functions and Diseases
| RBP Type | Canonical_RBPs |
| Diseases | Cancer |
| Drug | N.A. |
| Main interacting RNAs | mRNA |
| Moonlighting functions | N.A. |
| Localizations | P-body |
| BulkPerturb-seq | N.A. |
Description
| Ensembl ID | ENSG00000185650 | Gene ID | 677 | Accession | 1107 |
| Symbol | ZFP36L1 | Alias | BRF1;ERF1;cMG1;ERF-1;Berg36;TIS11B;RNF162B | Full Name | ZFP36 ring finger protein like 1 |
| Status | Confidence | Length | 8592 bases | Strand | Minus strand |
| Position | 14 : 68787662 - 68796253 | RNA binding domain | zf-CCCH , zf-CCCH_2 | ||
| Summary | This gene is a member of the TIS11 family of early response genes, which are induced by various agonists such as the phorbol ester TPA and the polypeptide mitogen EGF. This gene is well conserved across species and has a promoter that contains motifs seen in other early-response genes. The encoded protein contains a distinguishing putative zinc finger domain with a repeating cys-his motif. This putative nuclear transcription factor most likely functions in regulating the response to growth factors. Alternatively spliced transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Sep 2011] | ||||
RNA binding domains (RBDs)
| Protein | Domain | Pfam ID | E-value | Domain number |
|---|---|---|---|---|
| ENSP00000450600 | zf-CCCH | PF00642 | 8.7e-23 | 1 |
| ENSP00000450600 | zf-CCCH | PF00642 | 8.7e-23 | 2 |
| ENSP00000450600 | zf-CCCH_2 | PF14608 | 7e-06 | 3 |
| ENSP00000450600 | zf-CCCH_2 | PF14608 | 7e-06 | 4 |
| ENSP00000450784 | zf-CCCH | PF00642 | 1.4e-22 | 1 |
| ENSP00000450784 | zf-CCCH | PF00642 | 1.4e-22 | 2 |
| ENSP00000450784 | zf-CCCH_2 | PF14608 | 1.1e-05 | 3 |
| ENSP00000450784 | zf-CCCH_2 | PF14608 | 1.1e-05 | 4 |
| ENSP00000337386 | zf-CCCH | PF00642 | 8.3e-22 | 1 |
| ENSP00000337386 | zf-CCCH | PF00642 | 8.3e-22 | 2 |
| ENSP00000337386 | zf-CCCH_2 | PF14608 | 3.6e-05 | 3 |
| ENSP00000337386 | zf-CCCH_2 | PF14608 | 3.6e-05 | 4 |
| ENSP00000388402 | zf-CCCH | PF00642 | 8.3e-22 | 1 |
| ENSP00000388402 | zf-CCCH | PF00642 | 8.3e-22 | 2 |
| ENSP00000388402 | zf-CCCH_2 | PF14608 | 3.6e-05 | 3 |
| ENSP00000388402 | zf-CCCH_2 | PF14608 | 3.6e-05 | 4 |
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|---|---|---|---|
| 32687160 | Nuclear accumulation of ZFP36L1 is cell cycle-dependent and determined by a C-terminal serine-rich cluster. | Yuki Matsuura | 2020-11-01 | Journal of biochemistry |
| 36963393 | A miR-124-mediated post-transcriptional mechanism controlling the cell fate switch of astrocytes to induced neurons. | Elsa Papadimitriou | 2023-04-11 | Stem cell reports |
| 29993187 | Levamisole suppresses adipogenesis of aplastic anaemia-derived bone marrow mesenchymal stem cells through ZFP36L1-PPARGC1B axis. | Lu-Lu Liu | 2018-09-01 | Journal of cellular and molecular medicine |
| 34643435 | Zinc Finger Protein ZFP36L1 Inhibits Flavivirus Infection by both 5'-3' XRN1 and 3'-5' RNA-Exosome RNA Decay Pathways. | Han Chiu | 2022-01-12 | Journal of virology |
| 31999936 | Overexpression of microRNA-129-5p in glioblastoma inhibits cell proliferation, migration, and colony-forming ability by targeting ZFP36L1. | Xu Guo | 2020-11-02 | Bosnian journal of basic medical sciences |
| 22903503 | Identification of a novel flow-mediated gene expression signature in patients with bicuspid aortic valve. | Shohreh Maleki | 2013-01-01 | Journal of molecular medicine (Berlin, Germany) |
| 27102483 | RNA-binding proteins ZFP36L1 and ZFP36L2 promote cell quiescence. | Alison Galloway | 2016-04-22 | Science (New York, N.Y.) |
| 33427197 | Use of signals of positive and negative selection to distinguish cancer genes and passenger genes. | László Bányai | 2021-01-11 | eLife |
| 35611776 | ZFP36L1 Promotes Gastric Cancer Progression via Regulating JNK and p38 MAPK Signaling Pathways. | Kang Ding | 2023-01-01 | Recent patents on anti-cancer drug discovery |
| 32556261 | Zinc finger protein ZFP36L1 inhibits influenza A virus through translational repression by targeting HA, M and NS RNA transcripts. | Ren-Jye Lin | 2020-07-27 | Nucleic acids research |
| 31551365 | RNA-Binding Protein ZFP36L1 Suppresses Hypoxia and Cell-Cycle Signaling. | Xin-Yi Loh | 2020-01-15 | Cancer research |
| 30755618 | Whole-genome sequencing identifies ADGRG6 enhancer mutations and FRS2 duplications as angiogenesis-related drivers in bladder cancer. | Song Wu | 2019-02-12 | Nature communications |
| 36100056 | Silencing of RNA binding protein, ZFP36L1, promotes epithelial-mesenchymal transition in liver cancer cells by regulating transcription factor ZEB2. | Jian Chen | 2022-09-12 | Cellular signalling |
| 33711487 | Proteomic analysis of Laodelphax striatellus in response to Rice stripe virus infection reveal a potential role of ZFP36L1 in restriction of viral proliferation. | Hai-Jian Huang | 2021-05-15 | Journal of proteomics |
| 35642067 | The multifaceted role of the SASP in atherosclerosis: from mechanisms to therapeutic opportunities. | Yu Sun | 2022-05-31 | Cell & bioscience |
| 29257272 | High expression of FLT3 is a risk factor in leukemia. | Jie Cheng | 2018-02-01 | Molecular medicine reports |
| 30777893 | Quantification and discovery of sequence determinants of protein-per-mRNA amount in 29 human tissues. | Basak Eraslan | 2019-02-18 | Molecular systems biology |
| 36008402 | Regulation of neuroendocrine plasticity by the RNA-binding protein ZFP36L1. | Hsiao-Yun Chen | 2022-08-25 | Nature communications |
| 29868023 | The Interplay Between Innate-Like B Cells and Other Cell Types in Autoimmunity. | J Gregory Tsay | 2018-01-01 | Frontiers in immunology |
| 30622281 | RNA-binding protein ZFP36L1 regulates osteoarthritis by modulating members of the heat shock protein 70 family. | Young-Ok Son | 2019-01-08 | Nature communications |
| 37355988 | Understanding heterogeneity of human bone marrow plasma cell maturation and survival pathways by single-cell analyses. | Meixue Duan | 2023-07-25 | Cell reports |
| 35318269 | Killing SCLC: insights into how to target a shapeshifting tumor. | D Kate Sutherland | 2022-03-01 | Genes & development |
| 34540682 | Brain Microenvironment Heterogeneity: Potential Value for Brain Tumors. | Laura Álvaro-Espinosa | 2021-01-01 | Frontiers in oncology |
| 34295816 | The RNA-Binding Protein NELFE Promotes Gastric Cancer Growth and Metastasis Through E2F2. | Changyu Chen | 2021-01-01 | Frontiers in oncology |
| 33397444 | Comprehensive integrative profiling of upper tract urothelial carcinomas. | Xiaoping Su | 2021-01-04 | Genome biology |
| 36685529 | Comprehensive bulk and single-cell transcriptome profiling give useful insights into the characteristics of osteoarthritis associated synovial macrophages. | Shengyou Liao | 2022-01-01 | Frontiers in immunology |
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| ZFP36L1-204 | ENST00000555997 | 3094 | No protein | - |
| ZFP36L1-202 | ENST00000439696 | 3017 | 338aa | ENSP00000388402 |
| ZFP36L1-201 | ENST00000336440 | 4539 | 338aa | ENSP00000337386 |
| ZFP36L1-206 | ENST00000557086 | 727 | 207aa | ENSP00000450784 |
| ZFP36L1-205 | ENST00000557022 | 577 | 176aa | ENSP00000450600 |
| ZFP36L1-203 | ENST00000553375 | 633 | 189aa | ENSP00000452119 |
Phenotypes
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| 46502 | Platelet Function Tests | 1.2086900E-005 | - |
| 62247 | Age-Related Hearing Impairment 1 | 8E-9 | 24939585 |
| 66389 | Inflammatory Bowel Diseases | 1E-8 | 26192919 |
| 73285 | Diabetes Mellitus, Type 1 | 2E-12 | 19430480 |
| 83688 | Fibrinogen | 1E-8 | 23969696 |
| 83748 | Fibrinogen | 4E-13 | 28107422 |
| 85688 | Inflammatory Bowel Diseases | 3E-10 | 23128233 |
| 99278 | Crohn Disease | 1E-7 | 26192919 |
| 99791 | Fibrinogen | 8E-14 | 28107422 |
| 101561 | Fibrinogen | 2E-12 | 26561523 |
| 101901 | Celiac Disease | 4E-7 | 20190752 |
| 104772 | Multiple Sclerosis | 9E-12 | 21833088 |
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
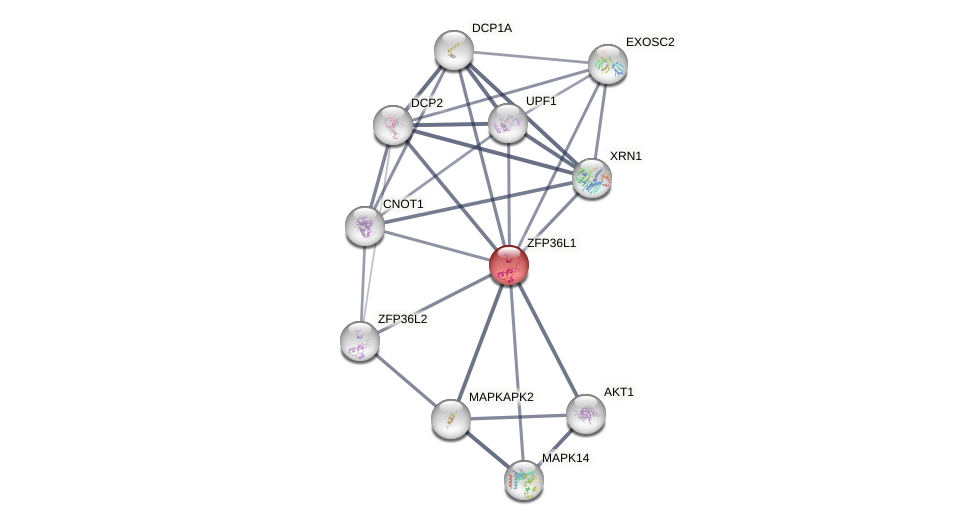
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000185650 | homo_sapiens | ENSP00000282388 | 46.7611 | 40.081 | homo_sapiens | ENSP00000388402 | 68.3432 | 58.5799 |
| ENSG00000185650 | homo_sapiens | ENSP00000469647 | 39.2638 | 32.8221 | homo_sapiens | ENSP00000388402 | 37.8698 | 31.6568 |
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 100 | 99.7041 | nomascus_leucogenys | ENSNLEP00000001225 | 100 | 99.7041 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.7041 | 99.7041 | pan_paniscus | ENSPPAP00000008436 | 99.7041 | 99.7041 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 100 | 99.7041 | pongo_abelii | ENSPPYP00000006744 | 100 | 99.7041 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.7041 | 99.7041 | pan_troglodytes | ENSPTRP00000061442 | 99.7041 | 99.7041 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 100 | 100 | gorilla_gorilla | ENSGGOP00000034936 | 100 | 100 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 93.1953 | 90.5325 | ornithorhynchus_anatinus | ENSOANP00000045066 | 93.75 | 91.0714 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 93.4911 | 91.4201 | sarcophilus_harrisii | ENSSHAP00000027971 | 76.699 | 75 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 96.1538 | 94.9704 | notamacropus_eugenii | ENSMEUP00000002797 | 96.1538 | 94.9704 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 76.0355 | 75.7396 | dasypus_novemcinctus | ENSDNOP00000003991 | 98.4674 | 98.0843 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.4083 | 99.4083 | callithrix_jacchus | ENSCJAP00000074036 | 99.4083 | 99.4083 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.4083 | 99.4083 | cercocebus_atys | ENSCATP00000041697 | 99.4083 | 99.4083 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.7041 | 99.7041 | macaca_fascicularis | ENSMFAP00000061805 | 99.7041 | 99.7041 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 95.5621 | 94.9704 | macaca_mulatta | ENSMMUP00000047605 | 93.8953 | 93.314 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 98.2249 | 97.929 | macaca_nemestrina | ENSMNEP00000017213 | 98.2249 | 97.929 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.7041 | 99.7041 | papio_anubis | ENSPANP00000017136 | 99.7041 | 99.7041 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.4083 | 99.4083 | mandrillus_leucophaeus | ENSMLEP00000040462 | 99.4083 | 99.4083 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.1124 | 99.1124 | tursiops_truncatus | ENSTTRP00000003257 | 99.1124 | 99.1124 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.1124 | 97.929 | vulpes_vulpes | ENSVVUP00000004531 | 99.1124 | 97.929 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 98.8166 | 98.5207 | mus_spretus | MGP_SPRETEiJ_P0034163 | 98.8166 | 98.5207 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 98.8166 | 98.8166 | equus_asinus | ENSEASP00005061881 | 98.8166 | 98.8166 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.1124 | 99.1124 | capra_hircus | ENSCHIP00000026339 | 99.1124 | 99.1124 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 98.8166 | 98.5207 | loxodonta_africana | ENSLAFP00000026015 | 98.8166 | 98.5207 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.1124 | 98.8166 | panthera_leo | ENSPLOP00000020614 | 99.1124 | 98.8166 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 89.0533 | 88.4615 | ailuropoda_melanoleuca | ENSAMEP00000027298 | 76.5903 | 76.0814 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 94.6746 | 94.6746 | bos_indicus_hybrid | ENSBIXP00005000977 | 99.0712 | 99.0712 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 42.0118 | 37.8698 | oryctolagus_cuniculus | ENSOCUP00000037215 | 69.6078 | 62.7451 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 98.8166 | 98.8166 | equus_caballus | ENSECAP00000016390 | 98.8166 | 98.8166 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.4083 | 98.2249 | canis_lupus_familiaris | ENSCAFP00845005268 | 99.4083 | 98.2249 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.4083 | 99.1124 | panthera_pardus | ENSPPRP00000024891 | 99.4083 | 99.1124 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.4083 | 99.4083 | marmota_marmota_marmota | ENSMMMP00000000490 | 99.4083 | 99.4083 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.4083 | 99.4083 | balaenoptera_musculus | ENSBMSP00010002595 | 99.4083 | 99.4083 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.1124 | 98.5207 | cavia_porcellus | ENSCPOP00000028707 | 99.1124 | 98.5207 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.4083 | 99.1124 | felis_catus | ENSFCAP00000023831 | 99.4083 | 99.1124 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.4083 | 99.1124 | ursus_americanus | ENSUAMP00000013365 | 99.4083 | 99.1124 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 87.574 | 85.503 | monodelphis_domestica | ENSMODP00000012674 | 94.2675 | 92.0382 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.1124 | 99.1124 | monodon_monoceros | ENSMMNP00015017839 | 99.1124 | 99.1124 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 97.6331 | 95.2663 | sus_scrofa | ENSSSCP00000060036 | 94.8276 | 92.5287 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.1124 | 99.1124 | microcebus_murinus | ENSMICP00000036424 | 99.1124 | 99.1124 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 98.8166 | 98.5207 | octodon_degus | ENSODEP00000019199 | 98.8166 | 98.5207 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 92.8994 | 92.0118 | jaculus_jaculus | ENSJJAP00000014905 | 91.5452 | 90.6706 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.1124 | 99.1124 | ovis_aries_rambouillet | ENSOARP00020004543 | 99.1124 | 99.1124 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.4083 | 99.1124 | ursus_maritimus | ENSUMAP00000013753 | 99.4083 | 99.1124 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 61.8343 | 58.284 | ochotona_princeps | ENSOPRP00000005500 | 66.3492 | 62.5397 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.1124 | 99.1124 | delphinapterus_leucas | ENSDLEP00000015850 | 99.1124 | 99.1124 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.1124 | 99.1124 | physeter_catodon | ENSPCTP00005021727 | 99.1124 | 99.1124 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 98.8166 | 98.5207 | mus_caroli | MGP_CAROLIEiJ_P0032655 | 98.8166 | 98.5207 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 98.8166 | 98.5207 | mus_musculus | ENSMUSP00000021552 | 98.8166 | 98.5207 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.4083 | 98.8166 | dipodomys_ordii | ENSDORP00000016057 | 99.4083 | 98.8166 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 92.6036 | 92.3077 | mustela_putorius_furo | ENSMPUP00000007225 | 98.7382 | 98.4227 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.4083 | 99.4083 | aotus_nancymaae | ENSANAP00000039478 | 99.4083 | 99.4083 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.7041 | 99.7041 | saimiri_boliviensis_boliviensis | ENSSBOP00000018085 | 99.7041 | 99.7041 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.7041 | 99.7041 | chlorocebus_sabaeus | ENSCSAP00000009290 | 99.7041 | 99.7041 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.1124 | 98.8166 | mesocricetus_auratus | ENSMAUP00000022734 | 99.1124 | 98.8166 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.1124 | 99.1124 | phocoena_sinus | ENSPSNP00000010674 | 99.1124 | 99.1124 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.4083 | 99.1124 | camelus_dromedarius | ENSCDRP00005004261 | 99.4083 | 99.1124 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 98.2249 | 98.2249 | carlito_syrichta | ENSTSYP00000033292 | 98.2249 | 98.2249 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 87.2781 | 86.6864 | panthera_tigris_altaica | ENSPTIP00000022298 | 98.0066 | 97.3422 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 98.5207 | 98.5207 | rattus_norvegicus | ENSRNOP00000075412 | 98.5207 | 98.5207 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.1124 | 99.1124 | prolemur_simus | ENSPSMP00000015879 | 99.1124 | 99.1124 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.1124 | 98.5207 | microtus_ochrogaster | ENSMOCP00000003983 | 99.1124 | 98.5207 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.1124 | 98.5207 | peromyscus_maniculatus_bairdii | ENSPEMP00000010350 | 99.1124 | 98.5207 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.1124 | 99.1124 | bos_mutus | ENSBMUP00000016850 | 99.1124 | 99.1124 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 61.2426 | 60.9467 | mus_spicilegus | ENSMSIP00000002015 | 98.5714 | 98.0952 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.1124 | 98.8166 | cricetulus_griseus_chok1gshd | ENSCGRP00001008685 | 99.1124 | 98.8166 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 93.1953 | 92.3077 | pteropus_vampyrus | ENSPVAP00000011017 | 98.7461 | 97.8056 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 95.5621 | 94.3787 | vombatus_ursinus | ENSVURP00010030553 | 95.5621 | 94.3787 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.1124 | 99.1124 | rhinolophus_ferrumequinum | ENSRFEP00010007255 | 99.1124 | 99.1124 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.1124 | 99.1124 | neovison_vison | ENSNVIP00000012135 | 99.1124 | 99.1124 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.1124 | 99.1124 | bos_taurus | ENSBTAP00000035611 | 99.1124 | 99.1124 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 98.8166 | 98.5207 | heterocephalus_glaber_female | ENSHGLP00000046854 | 98.8166 | 98.5207 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 94.9704 | 94.3787 | rhinopithecus_bieti | ENSRBIP00000033500 | 98.1651 | 97.5535 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.4083 | 98.2249 | canis_lupus_dingo | ENSCAFP00020007344 | 99.4083 | 98.2249 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.4083 | 99.4083 | sciurus_vulgaris | ENSSVLP00005016464 | 99.4083 | 99.4083 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 93.4911 | 91.4201 | phascolarctos_cinereus | ENSPCIP00000051178 | 80 | 78.2279 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.1124 | 99.1124 | procavia_capensis | ENSPCAP00000004972 | 99.1124 | 99.1124 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.1124 | 98.8166 | nannospalax_galili | ENSNGAP00000026854 | 99.1124 | 98.8166 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.1124 | 99.1124 | bos_grunniens | ENSBGRP00000010484 | 99.1124 | 99.1124 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 98.8166 | 98.2249 | chinchilla_lanigera | ENSCLAP00000005206 | 98.8166 | 98.2249 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.1124 | 98.8166 | cervus_hanglu_yarkandensis | ENSCHYP00000019991 | 99.1124 | 98.8166 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.4083 | 99.4083 | catagonus_wagneri | ENSCWAP00000029501 | 99.4083 | 99.4083 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.4083 | 99.4083 | ictidomys_tridecemlineatus | ENSSTOP00000008745 | 99.4083 | 99.4083 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 97.929 | 97.929 | otolemur_garnettii | ENSOGAP00000008964 | 97.929 | 97.929 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.7041 | 99.7041 | cebus_imitator | ENSCCAP00000001169 | 99.7041 | 99.7041 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 98.5207 | 98.5207 | urocitellus_parryii | ENSUPAP00010015403 | 97.3684 | 97.3684 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.1124 | 99.1124 | moschus_moschiferus | ENSMMSP00000019724 | 99.1124 | 99.1124 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.7041 | 99.7041 | rhinopithecus_roxellana | ENSRROP00000034070 | 99.7041 | 99.7041 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 98.8166 | 98.5207 | mus_pahari | MGP_PahariEiJ_P0082765 | 98.8166 | 98.5207 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 99.1124 | 98.8166 | propithecus_coquereli | ENSPCOP00000021052 | 99.1124 | 98.8166 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 25.7396 | 17.1598 | caenorhabditis_elegans | F52E1.1.1 | 32.9545 | 21.9697 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 21.3018 | 15.0888 | caenorhabditis_elegans | Y60A9.3.1 | 35.468 | 25.1232 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 23.6686 | 16.2722 | caenorhabditis_elegans | Y116A8C.17.1 | 39.0244 | 26.8293 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 23.6686 | 17.1598 | caenorhabditis_elegans | F38C2.5.1 | 43.0108 | 31.1828 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 20.7101 | 14.497 | caenorhabditis_elegans | C35D6.4.1 | 34.4828 | 24.1379 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 23.3728 | 16.2722 | caenorhabditis_elegans | Y116A8C.20.1 | 39.3035 | 27.3632 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 21.5976 | 15.0888 | caenorhabditis_elegans | Y116A8C.19.1 | 37.2449 | 26.0204 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 21.3018 | 14.7929 | caenorhabditis_elegans | F38C2.7.1 | 35.468 | 24.6305 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 22.7811 | 15.3846 | caenorhabditis_elegans | Y57G11C.25.1 | 38.6935 | 26.1307 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 26.0355 | 16.568 | caenorhabditis_elegans | W02A2.7.1 | 18.8034 | 11.9658 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 25.4438 | 15.9763 | caenorhabditis_elegans | AH6.5.1 | 18.4154 | 11.5632 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 28.4024 | 19.2308 | caenorhabditis_elegans | C09G9.6.1 | 23.5872 | 15.9705 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 30.1775 | 18.9349 | caenorhabditis_elegans | F32A11.6.1 | 27.7929 | 17.4387 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 28.6982 | 17.7515 | caenorhabditis_elegans | ZC513.6.1 | 24.6819 | 15.2672 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 41.1243 | 28.4024 | caenorhabditis_elegans | F38B7.1a.1 | 30.2174 | 20.8696 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 12.7219 | 7.39645 | caenorhabditis_elegans | F02E11.4.1 | 31.1594 | 18.1159 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 13.0178 | 7.98817 | caenorhabditis_elegans | Y110A2AL.1.1 | 30.9859 | 19.0141 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 10.0592 | 6.50888 | caenorhabditis_elegans | Y110A2AL.10.1 | 30.6306 | 19.8198 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 36.3905 | 24.5562 | caenorhabditis_elegans | T02E1.3b.1 | 18.1149 | 12.2239 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 74.8521 | 63.3136 | callorhinchus_milii | ENSCMIP00000038810 | 77.8462 | 65.8462 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 79.8817 | 70.4142 | latimeria_chalumnae | ENSLACP00000022123 | 78.0347 | 68.7861 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 76.0355 | 66.8639 | lepisosteus_oculatus | ENSLOCP00000017209 | 71.7877 | 63.1285 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 75.7396 | 65.6805 | danio_rerio | ENSDARP00000025205 | 68.4492 | 59.3583 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 72.7811 | 63.0177 | danio_rerio | ENSDARP00000011150 | 70.6897 | 61.2069 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 75.4438 | 65.9763 | carassius_auratus | ENSCARP00000076463 | 68.3646 | 59.7855 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 71.5976 | 60.9467 | carassius_auratus | ENSCARP00000102517 | 68.9459 | 58.6895 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 71.3018 | 60.355 | carassius_auratus | ENSCARP00000123681 | 62.4352 | 52.8497 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 71.0059 | 60.355 | carassius_auratus | ENSCARP00000023584 | 68.9655 | 58.6207 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 75.7396 | 66.2722 | carassius_auratus | ENSCARP00000119632 | 68.6327 | 60.0536 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 76.0355 | 66.568 | astyanax_mexicanus | ENSAMXP00000054122 | 68.7166 | 60.1604 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 67.7515 | 56.8047 | astyanax_mexicanus | ENSAMXP00000048370 | 67.3529 | 56.4706 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 75.1479 | 65.3846 | ictalurus_punctatus | ENSIPUP00000035317 | 68.2796 | 59.4086 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 73.3728 | 65.3846 | esox_lucius | ENSELUP00000007959 | 67.2087 | 59.8916 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 76.3314 | 66.8639 | esox_lucius | ENSELUP00000022557 | 65.3165 | 57.2152 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 60.6509 | 52.9586 | electrophorus_electricus | ENSEEEP00000013303 | 71.1806 | 62.1528 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 61.5385 | 51.4793 | electrophorus_electricus | ENSEEEP00000022793 | 59.2593 | 49.5727 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 72.7811 | 63.6095 | oncorhynchus_kisutch | ENSOKIP00005031461 | 62.1212 | 54.2929 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 75.4438 | 66.2722 | oncorhynchus_kisutch | ENSOKIP00005111140 | 58.0866 | 51.0251 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 74.5562 | 65.3846 | oncorhynchus_kisutch | ENSOKIP00005035859 | 65.7963 | 57.7024 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 68.9349 | 60.6509 | oncorhynchus_kisutch | ENSOKIP00005077417 | 67.341 | 59.2486 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 72.7811 | 63.9053 | oncorhynchus_mykiss | ENSOMYP00000065666 | 66.6667 | 58.5366 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 74.5562 | 66.2722 | oncorhynchus_mykiss | ENSOMYP00000062956 | 64.9485 | 57.732 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 75.4438 | 66.2722 | oncorhynchus_mykiss | ENSOMYP00000080509 | 61.0048 | 53.5885 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 69.5266 | 60.9467 | oncorhynchus_mykiss | ENSOMYP00000016033 | 68.314 | 59.8837 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 73.0769 | 64.7929 | salmo_salar | ENSSSAP00000144679 | 58.6698 | 52.019 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 74.8521 | 65.3846 | salmo_salar | ENSSSAP00000166676 | 64.0506 | 55.9494 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 74.5562 | 65.3846 | salmo_salar | ENSSSAP00000128653 | 64.6154 | 56.6667 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 69.5266 | 60.9467 | salmo_salar | ENSSSAP00000126800 | 59.949 | 52.551 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 74.5562 | 65.3846 | salmo_trutta | ENSSTUP00000110249 | 65.7963 | 57.7024 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 74.8521 | 65.0888 | salmo_trutta | ENSSTUP00000037482 | 54.5259 | 47.4138 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 69.2308 | 60.355 | salmo_trutta | ENSSTUP00000089523 | 67.8261 | 59.1304 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 73.3728 | 65.3846 | salmo_trutta | ENSSTUP00000114278 | 67.2087 | 59.8916 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 71.8935 | 63.6095 | gadus_morhua | ENSGMOP00000037639 | 55.7339 | 49.3119 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 72.4852 | 62.7219 | fundulus_heteroclitus | ENSFHEP00000021540 | 66.9399 | 57.9235 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 72.4852 | 62.7219 | poecilia_reticulata | ENSPREP00000019926 | 68.4358 | 59.2179 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 73.0769 | 62.7219 | xiphophorus_maculatus | ENSXMAP00000023749 | 66.5768 | 57.1429 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 74.2604 | 64.2012 | oryzias_latipes | ENSORLP00000044184 | 68.5792 | 59.2896 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 68.3432 | 57.3965 | cyclopterus_lumpus | ENSCLMP00005015260 | 64.7059 | 54.3417 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 71.3018 | 61.8343 | cyclopterus_lumpus | ENSCLMP00005023621 | 65.4891 | 56.7935 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 74.8521 | 62.7219 | oreochromis_niloticus | ENSONIP00000031092 | 68.0107 | 56.9892 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 74.8521 | 62.7219 | haplochromis_burtoni | ENSHBUP00000030796 | 68.0107 | 56.9892 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 74.8521 | 62.7219 | astatotilapia_calliptera | ENSACLP00000011348 | 68.0107 | 56.9892 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 76.0355 | 66.2722 | sparus_aurata | ENSSAUP00010068581 | 57.3661 | 50 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 67.1598 | 56.8047 | sparus_aurata | ENSSAUP00010006072 | 63.9437 | 54.0845 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 71.8935 | 61.5385 | lates_calcarifer | ENSLCAP00010025566 | 67.8771 | 58.1006 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 74.8521 | 64.497 | lates_calcarifer | ENSLCAP00010004541 | 59.9526 | 51.6588 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 81.3609 | 72.7811 | xenopus_tropicalis | ENSXETP00000037007 | 85.1393 | 76.161 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 91.1243 | 87.2781 | chrysemys_picta_bellii | ENSCPBP00000030561 | 91.9403 | 88.0597 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 92.0118 | 87.2781 | sphenodon_punctatus | ENSSPUP00000002641 | 92.5595 | 87.7976 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 92.8994 | 89.9408 | crocodylus_porosus | ENSCPRP00005025361 | 93.7313 | 90.7463 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 89.0533 | 84.6154 | laticauda_laticaudata | ENSLLTP00000008418 | 89.5833 | 85.119 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 89.3491 | 84.9112 | notechis_scutatus | ENSNSUP00000024473 | 89.881 | 85.4167 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 89.0533 | 84.6154 | pseudonaja_textilis | ENSPTXP00000017594 | 89.5833 | 85.119 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 91.1243 | 87.574 | anas_platyrhynchos_platyrhynchos | ENSAPLP00000032425 | 91.9403 | 88.3582 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 91.1243 | 87.2781 | gallus_gallus | ENSGALP00010029317 | 91.9403 | 88.0597 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 86.3905 | 82.5444 | meleagris_gallopavo | ENSMGAP00000020536 | 92.4051 | 88.2911 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 74.2604 | 70.1183 | serinus_canaria | ENSSCAP00000018209 | 85.0847 | 80.339 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 91.1243 | 87.8698 | parus_major | ENSPMJP00000020214 | 91.9403 | 88.6567 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 74.8521 | 62.7219 | neolamprologus_brichardi | ENSNBRP00000014004 | 68.0107 | 56.9892 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 89.645 | 85.2071 | naja_naja | ENSNNAP00000018531 | 90.1786 | 85.7143 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 90.5325 | 86.9822 | podarcis_muralis | ENSPMRP00000005307 | 91.0714 | 87.5 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 91.716 | 88.1657 | taeniopygia_guttata | ENSTGUP00000030630 | 92.5373 | 88.9552 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 76.3314 | 69.2308 | erpetoichthys_calabaricus | ENSECRP00000024323 | 72.8814 | 66.1017 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 74.2604 | 63.3136 | kryptolebias_marmoratus | ENSKMAP00000017636 | 68.7671 | 58.6301 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 90.5325 | 85.503 | salvator_merianae | ENSSMRP00000012231 | 91.0714 | 86.0119 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 74.2604 | 64.497 | oryzias_javanicus | ENSOJAP00000032684 | 68.2065 | 59.2391 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 73.0769 | 62.7219 | amphilophus_citrinellus | ENSACIP00000020992 | 71.8023 | 61.6279 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 74.8521 | 64.7929 | dicentrarchus_labrax | ENSDLAP00005029790 | 68.75 | 59.5109 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 68.9349 | 60.9467 | dicentrarchus_labrax | ENSDLAP00005079400 | 63.1436 | 55.8266 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 74.8521 | 64.497 | amphiprion_percula | ENSAPEP00000014362 | 68.0107 | 58.6021 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 74.2604 | 64.2012 | oryzias_sinensis | ENSOSIP00000020644 | 68.5792 | 59.2896 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 92.6036 | 89.0533 | pelodiscus_sinensis | ENSPSIP00000008952 | 93.4328 | 89.8507 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 91.1243 | 87.2781 | gopherus_evgoodei | ENSGEVP00005024186 | 92.2156 | 88.3234 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 90.8284 | 86.6864 | chelonoidis_abingdonii | ENSCABP00000023048 | 91.6418 | 87.4627 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 74.5562 | 64.497 | seriola_dumerili | ENSSDUP00000013318 | 67.7419 | 58.6021 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 68.3432 | 59.1716 | seriola_dumerili | ENSSDUP00000001269 | 64.8876 | 56.1798 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 75.1479 | 64.497 | cottoperca_gobio | ENSCGOP00000053327 | 68.6487 | 58.9189 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 71.3018 | 61.5385 | cottoperca_gobio | ENSCGOP00000008018 | 66.9444 | 57.7778 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 77.2189 | 74.5562 | anser_brachyrhynchus | ENSABRP00000017856 | 93.8849 | 90.6475 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 74.8521 | 64.2012 | gasterosteus_aculeatus | ENSGACP00000014895 | 67.2872 | 57.7128 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 74.2604 | 63.9053 | cynoglossus_semilaevis | ENSCSEP00000010975 | 67.1123 | 57.754 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 72.4852 | 62.1302 | poecilia_formosa | ENSPFOP00000025283 | 65.6836 | 56.3003 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 73.6686 | 65.0888 | myripristis_murdjan | ENSMMDP00005022005 | 70.339 | 62.1469 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 75.4438 | 65.6805 | myripristis_murdjan | ENSMMDP00005054649 | 68.3646 | 59.5174 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 76.0355 | 65.3846 | sander_lucioperca | ENSSLUP00000035042 | 68.9008 | 59.2493 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 69.5266 | 58.284 | sander_lucioperca | ENSSLUP00000035049 | 65.6425 | 55.0279 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 91.1243 | 87.574 | strigops_habroptila | ENSSHBP00005023194 | 91.9403 | 88.3582 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 74.8521 | 64.497 | amphiprion_ocellaris | ENSAOCP00000029131 | 68.0107 | 58.6021 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 91.1243 | 87.2781 | terrapene_carolina_triunguis | ENSTMTP00000011076 | 91.9403 | 88.0597 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 75.4438 | 65.0888 | hucho_hucho | ENSHHUP00000081316 | 64.2317 | 55.4156 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 72.7811 | 64.2012 | hucho_hucho | ENSHHUP00000056033 | 56.5517 | 49.8851 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 73.9645 | 65.0888 | hucho_hucho | ENSHHUP00000070082 | 65.1042 | 57.2917 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 69.2308 | 60.0592 | hucho_hucho | ENSHHUP00000052709 | 67.6301 | 58.6705 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 74.2604 | 65.3846 | betta_splendens | ENSBSLP00000054113 | 68.2065 | 60.0543 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 63.0177 | 52.6627 | betta_splendens | ENSBSLP00000027111 | 64.939 | 54.2683 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 66.568 | 57.6923 | pygocentrus_nattereri | ENSPNAP00000023121 | 66.9643 | 58.0357 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 76.0355 | 66.568 | pygocentrus_nattereri | ENSPNAP00000010204 | 68.7166 | 60.1604 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 74.8521 | 64.497 | anabas_testudineus | ENSATEP00000006431 | 68.0107 | 58.6021 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 70.1183 | 60.9467 | anabas_testudineus | ENSATEP00000020197 | 66.573 | 57.8652 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 74.5562 | 64.497 | seriola_lalandi_dorsalis | ENSSLDP00000027564 | 67.7419 | 58.6021 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 66.8639 | 57.9882 | seriola_lalandi_dorsalis | ENSSLDP00000013854 | 63.4831 | 55.0562 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 72.1893 | 62.7219 | labrus_bergylta | ENSLBEP00000010481 | 64.8936 | 56.383 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 74.8521 | 62.7219 | pundamilia_nyererei | ENSPNYP00000006431 | 68.1941 | 57.1429 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 74.5562 | 64.497 | mastacembelus_armatus | ENSMAMP00000000406 | 60.2871 | 52.1531 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 75.1479 | 64.7929 | stegastes_partitus | ENSSPAP00000013700 | 68.2796 | 58.871 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 73.0769 | 64.2012 | oncorhynchus_tshawytscha | ENSOTSP00005011189 | 66.9377 | 58.8076 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 73.0769 | 64.2012 | oncorhynchus_tshawytscha | ENSOTSP00005060381 | 66.9377 | 58.8076 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 74.5562 | 65.3846 | oncorhynchus_tshawytscha | ENSOTSP00005090775 | 65.7963 | 57.7024 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 69.2308 | 60.9467 | oncorhynchus_tshawytscha | ENSOTSP00005020040 | 67.6301 | 59.5376 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 75.4438 | 66.568 | oncorhynchus_tshawytscha | ENSOTSP00005013671 | 63.75 | 56.25 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 74.8521 | 64.497 | acanthochromis_polyacanthus | ENSAPOP00000017706 | 67.8284 | 58.445 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 90.8284 | 87.574 | aquila_chrysaetos_chrysaetos | ENSACCP00020006184 | 91.6418 | 88.3582 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 75.1479 | 63.6095 | nothobranchius_furzeri | ENSNFUP00015043308 | 68.8347 | 58.2656 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 73.0769 | 63.3136 | cyprinodon_variegatus | ENSCVAP00000024416 | 67.1196 | 58.1522 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 68.0473 | 58.5799 | denticeps_clupeoides | ENSDCDP00000003629 | 66.6667 | 57.3913 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 74.8521 | 65.0888 | larimichthys_crocea | ENSLCRP00005018021 | 68.0107 | 59.1398 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 64.2012 | 55.0296 | larimichthys_crocea | ENSLCRP00005037163 | 64.9701 | 55.6886 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 74.8521 | 64.497 | takifugu_rubripes | ENSTRUP00000080531 | 59.1121 | 50.9346 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 86.3905 | 83.1361 | ficedula_albicollis | ENSFALP00000015761 | 92.4051 | 88.924 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 39.3491 | 35.2071 | hippocampus_comes | ENSHCOP00000023871 | 66.1692 | 59.204 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 74.8521 | 66.2722 | hippocampus_comes | ENSHCOP00000011168 | 68.1941 | 60.3774 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 74.5562 | 65.0888 | cyprinus_carpio_carpio | ENSCCRP00000173572 | 61.6137 | 53.7897 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 72.1893 | 61.2426 | cyprinus_carpio_carpio | ENSCCRP00000158676 | 70.1149 | 59.4828 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 75.7396 | 66.568 | cyprinus_carpio_carpio | ENSCCRP00000166354 | 68.6327 | 60.3217 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 72.1893 | 61.8343 | cyprinus_carpio_carpio | ENSCCRP00000125697 | 69.5157 | 59.5442 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 91.1243 | 87.2781 | coturnix_japonica | ENSCJPP00005021954 | 91.9403 | 88.0597 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 72.1893 | 61.8343 | poecilia_latipinna | ENSPLAP00000004036 | 67.9666 | 58.2173 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 73.3728 | 62.1302 | sinocyclocheilus_grahami | ENSSGRP00000016051 | 70.6553 | 59.8291 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 75.4438 | 65.6805 | sinocyclocheilus_grahami | ENSSGRP00000006974 | 68.3646 | 59.5174 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 75.4438 | 66.2722 | sinocyclocheilus_grahami | ENSSGRP00000003625 | 68.5484 | 60.2151 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 74.8521 | 62.7219 | maylandia_zebra | ENSMZEP00005019554 | 68.0107 | 56.9892 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 73.3728 | 63.3136 | tetraodon_nigroviridis | ENSTNIP00000004746 | 72.7273 | 62.7566 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 75.1479 | 65.3846 | paramormyrops_kingsleyae | ENSPKIP00000027443 | 71.3483 | 62.0787 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 73.6686 | 65.9763 | paramormyrops_kingsleyae | ENSPKIP00000023808 | 68.9751 | 61.7729 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 72.7811 | 63.6095 | scleropages_formosus | ENSSFOP00015066881 | 66.8478 | 58.4239 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 76.3314 | 67.4556 | scleropages_formosus | ENSSFOP00015047204 | 70.8791 | 62.6374 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 58.284 | 48.5207 | scophthalmus_maximus | ENSSMAP00000029698 | 59.697 | 49.697 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 73.3728 | 63.6095 | scophthalmus_maximus | ENSSMAP00000053765 | 64.9215 | 56.2827 |
| ENSG00000185650 | homo_sapiens | ENSP00000388402 | 74.2604 | 64.497 | oryzias_melastigma | ENSOMEP00000016648 | 67.8378 | 58.9189 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0000165 | involved_in MAPK cascade | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0000288 | involved_in nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0000932 | located_in P-body | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0001570 | involved_in vasculogenesis | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0003342 | involved_in proepicardium development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0003677 | enables DNA binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0003723 | enables RNA binding | 1 | HDA | Homo_sapiens(9606) | Function |
| GO:0003729 | enables mRNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005634 | located_in nucleus | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005737 | located_in cytoplasm | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005829 | located_in cytosol | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0006397 | involved_in mRNA processing | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0006915 | involved_in apoptotic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0007507 | involved_in heart development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0008283 | involved_in cell population proliferation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0009611 | involved_in response to wounding | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0010468 | involved_in regulation of gene expression | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0010837 | involved_in regulation of keratinocyte proliferation | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0014065 | involved_in phosphatidylinositol 3-kinase signaling | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0021915 | involved_in neural tube development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0031086 | involved_in nuclear-transcribed mRNA catabolic process, deadenylation-independent decay | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0031440 | involved_in regulation of mRNA 3-end processing | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0032869 | involved_in cellular response to insulin stimulus | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0033077 | involved_in T cell differentiation in thymus | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0035264 | involved_in multicellular organism growth | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0035925 | enables mRNA 3-UTR AU-rich region binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0038066 | involved_in p38MAPK cascade | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0043488 | acts_upstream_of_or_within regulation of mRNA stability | 3 | IDA,IMP | Homo_sapiens(9606) | Process |
| GO:0043491 | involved_in protein kinase B signaling | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0044344 | involved_in cellular response to fibroblast growth factor stimulus | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0045577 | involved_in regulation of B cell differentiation | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0045600 | involved_in positive regulation of fat cell differentiation | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0045616 | involved_in regulation of keratinocyte differentiation | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0045647 | involved_in negative regulation of erythrocyte differentiation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0045657 | involved_in positive regulation of monocyte differentiation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0045661 | involved_in regulation of myoblast differentiation | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0046872 | enables metal ion binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0048382 | involved_in mesendoderm development | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0051028 | involved_in mRNA transport | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0060710 | involved_in chorio-allantoic fusion | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0061158 | involved_in 3-UTR-mediated mRNA destabilization | 2 | IDA,IMP | Homo_sapiens(9606) | Process |
| GO:0070371 | involved_in ERK1 and ERK2 cascade | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0071320 | involved_in cellular response to cAMP | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0071356 | involved_in cellular response to tumor necrosis factor | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0071364 | involved_in cellular response to epidermal growth factor stimulus | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0071375 | involved_in cellular response to peptide hormone stimulus | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0071385 | involved_in cellular response to glucocorticoid stimulus | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0071456 | involved_in cellular response to hypoxia | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0071472 | involved_in cellular response to salt stress | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0071560 | involved_in cellular response to transforming growth factor beta stimulus | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0071889 | enables 14-3-3 protein binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0072091 | involved_in regulation of stem cell proliferation | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0097403 | involved_in cellular response to raffinose | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:1900153 | involved_in positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:1901991 | involved_in negative regulation of mitotic cell cycle phase transition | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:1902172 | involved_in regulation of keratinocyte apoptotic process | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:1904582 | involved_in positive regulation of intracellular mRNA localization | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:1990904 | part_of ribonucleoprotein complex | 1 | IDA | Homo_sapiens(9606) | Component |