Welcome to
RBP World!
Functions and Diseases
| RBP Type | Canonical_RBPs |
| Diseases | Cancer Disease |
| Drug | N.A. |
| Main interacting RNAs | mRNA |
| Moonlighting functions | TF |
| Localizations | N.A. |
| BulkPerturb-seq | DataSet_02_64 |
Description
| Ensembl ID | ENSG00000169057 | Gene ID | 4204 | Accession | 6990 |
| Symbol | MECP2 | Alias | RS;RTS;RTT;PPMX;MRX16;MRX79;MRXSL;AUTSX3;MRXS13 | Full Name | methyl-CpG binding protein 2 |
| Status | Confidence | Length | 76165 bases | Strand | Minus strand |
| Position | X : 154021573 - 154097737 | RNA binding domain | MBD | ||
| Summary | DNA methylation is the major modification of eukaryotic genomes and plays an essential role in mammalian development. Human proteins MECP2, MBD1, MBD2, MBD3, and MBD4 comprise a family of nuclear proteins related by the presence in each of a methyl-CpG binding domain (MBD). Each of these proteins, with the exception of MBD3, is capable of binding specifically to methylated DNA. MECP2, MBD1 and MBD2 can also repress transcription from methylated gene promoters. In contrast to other MBD family members, MECP2 is X-linked and subject to X inactivation. MECP2 is dispensible in stem cells, but is essential for embryonic development. MECP2 gene mutations are the cause of most cases of Rett syndrome, a progressive neurologic developmental disorder and one of the most common causes of cognitive disability in females. Alternative splicing results in multiple transcript variants encoding different isoforms. [provided by RefSeq, Oct 2015] | ||||
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|---|---|---|---|
| 30703685 | Alternative splicing of neuronal genes: new mechanisms and new therapies. | Diane Lipscombe | 2019-08-01 | Current opinion in neurobiology |
| 37182097 | Potential role of N6-adenine DNA methylation in alternative splicing and endosymbiosis in Paramecium bursaria. | Bo Pan | 2023-05-19 | iScience |
| 28594324 | MeCP2 regulates Tet1-catalyzed demethylation, CTCF binding, and learning-dependent alternative splicing of the BDNF gene in Turtle. | Zhaoqing Zheng | 2017-06-08 | eLife |
| 33552148 | MeCP2: The Genetic Driver of Rett Syndrome Epigenetics. | V Katrina Good | 2021-01-01 | Frontiers in genetics |
| 36443876 | Gene body methylation in cancer: molecular mechanisms and clinical applications. | Qi Wang | 2022-11-28 | Clinical epigenetics |
| 18820302 | MECP2 genomic structure and function: insights from ENCODE. | Jasmine Singh | 2008-11-01 | Nucleic acids research |
| 22344696 | Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. | E Jeffrey Squires | 2012-06-01 | Nucleic acids research |
| 33585981 | Ectopic expression of pericentric HSATII RNA results in nuclear RNA accumulation, MeCP2 recruitment, and cell division defects. | C Catherine Landers | 2021-03-01 | Chromosoma |
| 28329686 | Demethylated HSATII DNA and HSATII RNA Foci Sequester PRC1 and MeCP2 into Cancer-Specific Nuclear Bodies. | L Lisa Hall | 2017-03-21 | Cell reports |
| 36857328 | Rett syndrome severity estimation with the BioStamp nPoint using interactions between heart rate variability and body movement. | Byappanahalli Pradyumna Suresha | 2023-01-01 | PloS one |
| 21070191 | A brain-derived MeCP2 complex supports a role for MeCP2 in RNA processing. | W Steven Long | 2011-10-01 | Bioscience reports |
| 25663474 | CXCR3 in carcinoma progression. | Bo Ma | 2015-07-01 | Histology and histopathology |
| 32681172 | Regulation, diversity and function of MECP2 exon and 3'UTR isoforms. | Carvalho Deivid Rodrigues | 2020-09-30 | Human molecular genetics |
| 30781346 | Antiglycative Activity and RAGE Expression in Rett Syndrome. | Valeria Cordone | 2019-02-15 | Cells |
| 34048843 | Proteomic and transcriptional changes associated with MeCP2 dysfunction reveal nodes for therapeutic intervention in Rett syndrome. | Ketan Marballi | 2021-09-01 | Neurochemistry international |
| 31329578 | Aberrant DNA methylation defines isoform usage in cancer, with functional implications. | Yun-Ching Chen | 2019-07-01 | PLoS computational biology |
| 23457515 | Reduced syncytin-1 expression levels in placental syndromes correlates with epigenetic hypermethylation of the ERVW-1 promoter region. | Matthias Ruebner | 2013-01-01 | PloS one |
| 29637074 | Gaussian-Based Smooth Dielectric Function: A Surface-Free Approach for Modeling Macromolecular Binding in Solvents. | Arghya Chakravorty | 2018-01-01 | Frontiers in molecular biosciences |
| 36323715 | Bioinformatics analysis of prognostic value and immunological role of MeCP2 in pan-cancer. | Yanfeng Wang | 2022-11-02 | Scientific reports |
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| MECP2-201 | ENST00000303391 | 10467 | 486aa | ENSP00000301948 |
| MECP2-205 | ENST00000453960 | 10343 | 498aa | ENSP00000395535 |
| MECP2-216 | ENST00000628176 | 1712 | 172aa | ENSP00000486978 |
| MECP2-203 | ENST00000407218 | 1174 | 184aa | ENSP00000384865 |
| MECP2-223 | ENST00000637917 | 159 | 53aa | ENSP00000489847 |
| MECP2-209 | ENST00000486506 | 2907 | No protein | - |
| MECP2-202 | ENST00000369957 | 784 | 37aa | ENSP00000358973 |
| MECP2-211 | ENST00000496908 | 342 | No protein | - |
| MECP2-219 | ENST00000631210 | 490 | No protein | - |
| MECP2-208 | ENST00000481807 | 436 | No protein | - |
| MECP2-210 | ENST00000488293 | 1198 | No protein | - |
| MECP2-206 | ENST00000460227 | 1298 | No protein | - |
| MECP2-207 | ENST00000463644 | 1088 | No protein | - |
| MECP2-204 | ENST00000415944 | 413 | 49aa | ENSP00000416267 |
| MECP2-213 | ENST00000625300 | 350 | No protein | - |
| MECP2-214 | ENST00000626422 | 835 | No protein | - |
| MECP2-218 | ENST00000630151 | 322 | 41aa | ENSP00000486089 |
| MECP2-228 | ENST00000676382 | 313 | No protein | - |
| MECP2-212 | ENST00000611468 | 366 | No protein | - |
| MECP2-221 | ENST00000637533 | 140 | No protein | - |
| MECP2-226 | ENST00000675526 | 769 | 61aa | ENSP00000501710 |
| MECP2-229 | ENST00000700484 | 197 | No protein | - |
| MECP2-222 | ENST00000637791 | 101 | No protein | - |
| MECP2-227 | ENST00000675841 | 1201 | No protein | - |
| MECP2-224 | ENST00000638041 | 100 | No protein | - |
| MECP2-225 | ENST00000674996 | 397 | 58aa | ENSP00000502832 |
| MECP2-215 | ENST00000627864 | 581 | No protein | - |
| MECP2-220 | ENST00000637467 | 101 | No protein | - |
| MECP2-217 | ENST00000629277 | 2558 | No protein | - |
Phenotypes
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| 49137 | Child Development Disorders, Pervasive | 4.8814100E-005 | - |
| 49138 | Child Development Disorders, Pervasive | 5.9476100E-005 | - |
| 49173 | Child Development Disorders, Pervasive | 4.8814100E-005 | - |
| 50579 | Child Development Disorders, Pervasive | 4.6750400E-006 | - |
| 50580 | Child Development Disorders, Pervasive | 2.5642600E-005 | - |
| 50581 | Child Development Disorders, Pervasive | 3.4092400E-006 | - |
| 50582 | Child Development Disorders, Pervasive | 2.8884900E-006 | - |
| 50583 | Child Development Disorders, Pervasive | 1.0967600E-006 | - |
| 50586 | Child Development Disorders, Pervasive | 1.0401500E-005 | - |
| 50591 | Child Development Disorders, Pervasive | 4.7229200E-006 | - |
| 50623 | Child Development Disorders, Pervasive | 4.6750400E-006 | - |
| 50624 | Child Development Disorders, Pervasive | 2.5642600E-005 | - |
| 50625 | Child Development Disorders, Pervasive | 3.4092400E-006 | - |
| 50626 | Child Development Disorders, Pervasive | 2.8884900E-006 | - |
| 50627 | Child Development Disorders, Pervasive | 1.0967600E-006 | - |
| 50632 | Child Development Disorders, Pervasive | 1.0401500E-005 | - |
| 50635 | Child Development Disorders, Pervasive | 4.7229200E-006 | - |
| 51310 | Child Development Disorders, Pervasive | 3.3854300E-005 | - |
| 51311 | Child Development Disorders, Pervasive | 2.9114700E-005 | - |
| 51312 | Child Development Disorders, Pervasive | 1.3535900E-005 | - |
| 51315 | Child Development Disorders, Pervasive | 3.5188000E-005 | - |
| 51347 | Child Development Disorders, Pervasive | 3.3854300E-005 | - |
| 51348 | Child Development Disorders, Pervasive | 2.9114700E-005 | - |
| 51349 | Child Development Disorders, Pervasive | 1.3535900E-005 | - |
| 51358 | Child Development Disorders, Pervasive | 3.5188000E-005 | - |
| 72282 | Lupus Erythematosus, Systemic | 2E-15 | 26502338 |
| 76835 | Arthritis, Rheumatoid | 5E-16 | 24390342 |
| 77284 | Arthritis, Rheumatoid | 3E-12 | 24390342 |
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
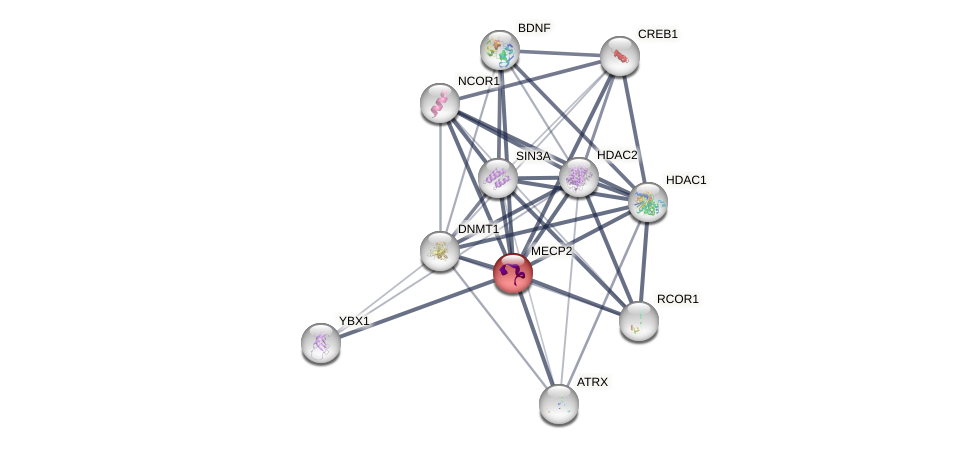
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000169057 | homo_sapiens | ENSP00000394080 | 29.2683 | 16.899 | homo_sapiens | ENSP00000395535 | 33.7349 | 19.4779 |
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 31.3253 | 31.3253 | nomascus_leucogenys | ENSNLEP00000036087 | 100 | 100 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 94.9799 | 94.7791 | pan_paniscus | ENSPPAP00000021674 | 95.9432 | 95.7404 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 100 | 100 | pongo_abelii | ENSPPYP00000033017 | 100 | 100 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 100 | 100 | pan_troglodytes | ENSPTRP00000081799 | 100 | 100 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 100 | 100 | gorilla_gorilla | ENSGGOP00000044548 | 100 | 100 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 30.7229 | 29.5181 | ornithorhynchus_anatinus | ENSOANP00000041870 | 93.2927 | 89.6341 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 89.9598 | 85.1406 | sarcophilus_harrisii | ENSSHAP00000009065 | 89.4212 | 84.6307 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 90.3614 | 85.1406 | notamacropus_eugenii | ENSMEUP00000002523 | 90 | 84.8 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 83.7349 | 81.1245 | dasypus_novemcinctus | ENSDNOP00000010402 | 91.6484 | 88.7912 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 69.8795 | 68.2731 | erinaceus_europaeus | ENSEEUP00000002809 | 74.6781 | 72.9614 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 51.2048 | 50.8032 | erinaceus_europaeus | ENSEEUP00000013705 | 95.5056 | 94.7566 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 87.5502 | 84.3373 | echinops_telfairi | ENSETEP00000007980 | 88.7984 | 85.5397 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 99.7992 | 99.5984 | callithrix_jacchus | ENSCJAP00000026578 | 100 | 99.7988 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 97.7912 | 96.988 | cercocebus_atys | ENSCATP00000005272 | 97.2056 | 96.4072 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 100 | 99.7992 | macaca_fascicularis | ENSMFAP00000020601 | 100 | 99.7992 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 100 | 99.7992 | macaca_mulatta | ENSMMUP00000055953 | 100 | 99.7992 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 100 | 99.7992 | macaca_nemestrina | ENSMNEP00000003766 | 100 | 99.7992 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 99.7992 | 99.5984 | papio_anubis | ENSPANP00000044243 | 99.7992 | 99.5984 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 95.1807 | 94.3775 | mandrillus_leucophaeus | ENSMLEP00000017207 | 95.1807 | 94.3775 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 96.7871 | 95.5823 | tursiops_truncatus | ENSTTRP00000004362 | 96.7871 | 95.5823 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 94.1767 | 93.3735 | vulpes_vulpes | ENSVVUP00000041582 | 92.5049 | 91.716 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 95.7831 | 94.9799 | mus_spretus | MGP_SPRETEiJ_P0095577 | 95.0199 | 94.2231 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 88.5542 | 87.751 | equus_asinus | ENSEASP00005028224 | 88.3768 | 87.5751 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 92.7711 | 91.3655 | capra_hircus | ENSCHIP00000026916 | 96.0499 | 94.5946 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 90.1606 | 88.755 | loxodonta_africana | ENSLAFP00000013674 | 95.7356 | 94.2431 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 97.3896 | 96.5863 | panthera_leo | ENSPLOP00000015531 | 97.3896 | 96.5863 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 97.3896 | 96.3855 | ailuropoda_melanoleuca | ENSAMEP00000032992 | 97.3896 | 96.3855 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 96.988 | 95.5823 | bos_indicus_hybrid | ENSBIXP00005032578 | 96.988 | 95.5823 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 94.7791 | 94.1767 | oryctolagus_cuniculus | ENSOCUP00000048554 | 97.3196 | 96.701 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 97.992 | 96.988 | equus_caballus | ENSECAP00000048042 | 97.992 | 96.988 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 93.9759 | 92.7711 | canis_lupus_familiaris | ENSCAFP00845037701 | 93.9759 | 92.7711 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 97.3896 | 96.5863 | panthera_pardus | ENSPPRP00000010444 | 97.3896 | 96.5863 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 98.996 | 98.5944 | marmota_marmota_marmota | ENSMMMP00000011182 | 98.7976 | 98.3968 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 94.5783 | 92.3695 | balaenoptera_musculus | ENSBMSP00010027488 | 92.8994 | 90.7298 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 92.1687 | 90.3614 | cavia_porcellus | ENSCPOP00000011316 | 91.0714 | 89.2857 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 97.3896 | 96.5863 | felis_catus | ENSFCAP00000049542 | 97.3896 | 96.5863 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 73.0924 | 70.6827 | ursus_americanus | ENSUAMP00000026386 | 82.7273 | 80 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 87.9518 | 82.9317 | monodelphis_domestica | ENSMODP00000014588 | 81.5642 | 76.9088 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 96.5863 | 95.3815 | monodon_monoceros | ENSMMNP00015017618 | 96.5863 | 95.3815 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 89.1566 | 87.3494 | sus_scrofa | ENSSSCP00000013606 | 96.3124 | 94.3601 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 98.5944 | 98.1928 | microcebus_murinus | ENSMICP00000004687 | 98.004 | 97.6048 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 91.5663 | 89.9598 | octodon_degus | ENSODEP00000006018 | 90.8367 | 89.243 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 97.3896 | 96.1847 | jaculus_jaculus | ENSJJAP00000022396 | 97.3896 | 96.1847 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 97.1888 | 95.7831 | ovis_aries_rambouillet | ENSOARP00020028463 | 97.1888 | 95.7831 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 31.1245 | 29.3173 | ursus_maritimus | ENSUMAP00000028560 | 90.1163 | 84.8837 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 93.1727 | 91.9679 | delphinapterus_leucas | ENSDLEP00000014805 | 95.4733 | 94.2387 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 95.9839 | 94.5783 | physeter_catodon | ENSPCTP00005030942 | 95.9839 | 94.5783 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 96.1847 | 95.3815 | mus_caroli | MGP_CAROLIEiJ_P0091823 | 95.6088 | 94.8104 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 95.9839 | 94.7791 | mus_musculus | ENSMUSP00000033770 | 95.4092 | 94.2116 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 97.5904 | 96.7871 | dipodomys_ordii | ENSDORP00000026159 | 97.2 | 96.4 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 91.9679 | 89.9598 | mustela_putorius_furo | ENSMPUP00000008999 | 94.6281 | 92.562 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 93.9759 | 92.9719 | aotus_nancymaae | ENSANAP00000003548 | 93.7876 | 92.7856 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 94.5783 | 93.9759 | saimiri_boliviensis_boliviensis | ENSSBOP00000016742 | 95.5375 | 94.929 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 95.7831 | 95.5823 | chlorocebus_sabaeus | ENSCSAP00000003084 | 99.5825 | 99.3737 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 96.5863 | 95.5823 | mesocricetus_auratus | ENSMAUP00000020085 | 95.4365 | 94.4444 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 96.5863 | 95.3815 | phocoena_sinus | ENSPSNP00000030261 | 96.5863 | 95.3815 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 59.0361 | 56.2249 | camelus_dromedarius | ENSCDRP00005016221 | 90.4615 | 86.1538 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 94.5783 | 93.7751 | carlito_syrichta | ENSTSYP00000018942 | 92.3529 | 91.5686 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 40.5622 | 39.5582 | panthera_tigris_altaica | ENSPTIP00000000938 | 92.6606 | 90.367 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 96.5863 | 95.1807 | rattus_norvegicus | ENSRNOP00000094658 | 92.5 | 91.1538 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 94.3775 | 93.7751 | prolemur_simus | ENSPSMP00000032861 | 94.3775 | 93.7751 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 96.3855 | 95.3815 | microtus_ochrogaster | ENSMOCP00000001709 | 94.6746 | 93.6884 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 89.5582 | 86.9478 | sorex_araneus | ENSSARP00000011669 | 95.0959 | 92.3241 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 96.3855 | 95.3815 | peromyscus_maniculatus_bairdii | ENSPEMP00000015659 | 95.0495 | 94.0594 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 59.4378 | 56.0241 | bos_mutus | ENSBMUP00000019393 | 91.358 | 86.1111 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 96.1847 | 95.3815 | mus_spicilegus | ENSMSIP00000029021 | 95.4183 | 94.6215 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 97.3896 | 96.1847 | cricetulus_griseus_chok1gshd | ENSCGRP00001017675 | 96.2302 | 95.0397 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 71.2851 | 70.4819 | pteropus_vampyrus | ENSPVAP00000000442 | 71.5726 | 70.7661 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 92.3695 | 91.9679 | myotis_lucifugus | ENSMLUP00000013005 | 95.6341 | 95.2183 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 90.3614 | 85.5422 | vombatus_ursinus | ENSVURP00010027906 | 89.8204 | 85.0299 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 76.9076 | 70.0803 | vombatus_ursinus | ENSVURP00010019553 | 80.4622 | 73.3193 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 97.7912 | 97.1888 | rhinolophus_ferrumequinum | ENSRFEP00010006307 | 97.7912 | 97.1888 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 92.3695 | 90.9639 | neovison_vison | ENSNVIP00000031024 | 94.8454 | 93.4021 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 96.988 | 95.5823 | bos_taurus | ENSBTAP00000054579 | 96.988 | 95.5823 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 74.0964 | 71.4859 | heterocephalus_glaber_female | ENSHGLP00000001139 | 88.4892 | 85.3717 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 94.9799 | 94.1767 | rhinopithecus_bieti | ENSRBIP00000038982 | 94.9799 | 94.1767 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 93.9759 | 92.7711 | canis_lupus_dingo | ENSCAFP00020028144 | 93.9759 | 92.7711 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 98.996 | 98.5944 | sciurus_vulgaris | ENSSVLP00005022622 | 98.996 | 98.5944 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 90.7631 | 85.9438 | phascolarctos_cinereus | ENSPCIP00000021603 | 89.8608 | 85.0895 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 94.9799 | 92.5703 | procavia_capensis | ENSPCAP00000010978 | 95.749 | 93.3198 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 97.5904 | 96.5863 | nannospalax_galili | ENSNGAP00000006236 | 97.3948 | 96.3928 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 96.3855 | 94.9799 | bos_grunniens | ENSBGRP00000037109 | 96.7742 | 95.3629 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 95.9839 | 94.1767 | chinchilla_lanigera | ENSCLAP00000002277 | 96.5657 | 94.7475 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 93.7751 | 92.1687 | cervus_hanglu_yarkandensis | ENSCHYP00000037302 | 96.0905 | 94.4444 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 88.5542 | 87.3494 | catagonus_wagneri | ENSCWAP00000011738 | 91.684 | 90.4366 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 95.5823 | 95.1807 | ictidomys_tridecemlineatus | ENSSTOP00000026241 | 97.9424 | 97.5309 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 95.7831 | 95.1807 | otolemur_garnettii | ENSOGAP00000013532 | 98.1481 | 97.5309 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 98.7952 | 97.5904 | cebus_imitator | ENSCCAP00000039146 | 98.7952 | 97.5904 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 98.996 | 98.5944 | urocitellus_parryii | ENSUPAP00010013535 | 98.996 | 98.5944 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 68.0723 | 64.8594 | moschus_moschiferus | ENSMMSP00000020804 | 90.6417 | 86.3636 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 96.7871 | 95.1807 | moschus_moschiferus | ENSMMSP00000020375 | 96.5932 | 94.99 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 94.9799 | 94.1767 | rhinopithecus_roxellana | ENSRROP00000043242 | 94.9799 | 94.1767 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 96.1847 | 95.3815 | mus_pahari | MGP_PahariEiJ_P0089450 | 95.0397 | 94.246 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 94.1767 | 93.3735 | propithecus_coquereli | ENSPCOP00000025258 | 93.8 | 93 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 18.2731 | 15.261 | petromyzon_marinus | ENSPMAP00000008581 | 69.4657 | 58.0153 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 15.4618 | 12.8514 | eptatretus_burgeri | ENSEBUP00000011869 | 60.6299 | 50.3937 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 51.004 | 42.1687 | callorhinchus_milii | ENSCMIP00000007913 | 63.1841 | 52.2388 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 75.3012 | 68.6747 | latimeria_chalumnae | ENSLACP00000012808 | 81.6993 | 74.5098 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 66.6667 | 59.0361 | lepisosteus_oculatus | ENSLOCP00000018131 | 66.5331 | 58.9178 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 50 | 39.759 | clupea_harengus | ENSCHAP00000055178 | 50.1006 | 39.839 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 60.0402 | 51.6064 | danio_rerio | ENSDARP00000109399 | 57.0611 | 49.0458 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 61.6466 | 52.2088 | carassius_auratus | ENSCARP00000119752 | 56.4338 | 47.7941 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 62.0482 | 52.4096 | carassius_auratus | ENSCARP00000110962 | 56.8015 | 47.9779 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 61.4458 | 51.8072 | carassius_auratus | ENSCARP00000031298 | 56.3536 | 47.5138 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 61.245 | 51.8072 | astyanax_mexicanus | ENSAMXP00000046961 | 49.8366 | 42.1569 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 61.6466 | 52.2088 | ictalurus_punctatus | ENSIPUP00000019201 | 57.5985 | 48.7805 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 59.4378 | 51.004 | esox_lucius | ENSELUP00000039027 | 48.2871 | 41.4356 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 57.8313 | 49.1968 | electrophorus_electricus | ENSEEEP00000006746 | 56.25 | 47.8516 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 63.6546 | 54.4177 | oncorhynchus_kisutch | ENSOKIP00005014325 | 43.3653 | 37.0725 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 62.6506 | 52.6104 | oncorhynchus_kisutch | ENSOKIP00005094005 | 38.5661 | 32.3857 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 58.2329 | 49.5984 | oncorhynchus_mykiss | ENSOMYP00000133305 | 34.6062 | 29.4749 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 60.6426 | 52.008 | oncorhynchus_mykiss | ENSOMYP00000009711 | 42.6554 | 36.5819 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 58.6345 | 49.1968 | salmo_salar | ENSSSAP00000162991 | 48.505 | 40.6977 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 62.4498 | 53.2129 | salmo_salar | ENSSSAP00000044090 | 46.1424 | 39.3175 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 63.4538 | 54.4177 | salmo_trutta | ENSSTUP00000023286 | 48.0243 | 41.1854 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 61.8474 | 51.6064 | salmo_trutta | ENSSTUP00000015807 | 34.5291 | 28.8117 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 54.6185 | 43.3735 | gadus_morhua | ENSGMOP00000063658 | 37.5172 | 29.7931 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 58.6345 | 46.988 | fundulus_heteroclitus | ENSFHEP00000031715 | 33.1442 | 26.5607 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 63.253 | 51.2048 | poecilia_reticulata | ENSPREP00000018569 | 32.0774 | 25.9674 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 62.6506 | 50.8032 | xiphophorus_maculatus | ENSXMAP00000037486 | 26.4407 | 21.4407 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 58.6345 | 45.7831 | oryzias_latipes | ENSORLP00000045247 | 29.0837 | 22.7092 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 53.8153 | 45.3815 | cyclopterus_lumpus | ENSCLMP00005000860 | 51.4395 | 43.3781 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 65.0602 | 53.012 | oreochromis_niloticus | ENSONIP00000029287 | 30.1115 | 24.5353 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 22.8916 | 18.6747 | haplochromis_burtoni | ENSHBUP00000033650 | 64.4068 | 52.5424 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 65.4618 | 53.2129 | astatotilapia_calliptera | ENSACLP00000020135 | 30.7838 | 25.0236 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 62.8514 | 51.6064 | lates_calcarifer | ENSLCAP00010037882 | 30.3883 | 24.9515 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 76.7068 | 67.8715 | xenopus_tropicalis | ENSXETP00000003337 | 81.9743 | 72.5322 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 33.5341 | 29.3173 | chrysemys_picta_bellii | ENSCPBP00000008868 | 71.9828 | 62.931 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 24.6988 | 22.2892 | laticauda_laticaudata | ENSLLTP00000010117 | 86.6197 | 78.169 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 75.9036 | 69.0763 | notechis_scutatus | ENSNSUP00000020477 | 77.7778 | 70.7819 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 77.3092 | 70.4819 | pseudonaja_textilis | ENSPTXP00000006762 | 81.0526 | 73.8947 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 59.0361 | 51.6064 | gallus_gallus | ENSGALP00010002474 | 41.9401 | 36.6619 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 77.9116 | 70.6827 | anolis_carolinensis | ENSACAP00000012271 | 75.0484 | 68.0851 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 56.0241 | 46.1847 | neolamprologus_brichardi | ENSNBRP00000010858 | 53.0418 | 43.7262 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 77.7108 | 70.8835 | naja_naja | ENSNNAP00000029167 | 81.4737 | 74.3158 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 28.7149 | 26.9076 | podarcis_muralis | ENSPMRP00000033560 | 90.5063 | 84.8101 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 57.0281 | 51.8072 | taeniopygia_guttata | ENSTGUP00000021498 | 74.3456 | 67.5393 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 59.6386 | 51.4056 | taeniopygia_guttata | ENSTGUP00000031243 | 43.2945 | 37.3178 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 21.8876 | 18.4739 | erpetoichthys_calabaricus | ENSECRP00000027450 | 66.0606 | 55.7576 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 62.0482 | 51.004 | kryptolebias_marmoratus | ENSKMAP00000020135 | 29.2891 | 24.0758 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 77.1084 | 69.2771 | salvator_merianae | ENSSMRP00000002126 | 81.3559 | 73.0932 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 58.6345 | 46.988 | oryzias_javanicus | ENSOJAP00000003809 | 30.6401 | 24.554 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 63.6546 | 52.8112 | dicentrarchus_labrax | ENSDLAP00005077477 | 29.2976 | 24.3068 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 50.6024 | 42.9719 | amphiprion_percula | ENSAPEP00000033846 | 38.8889 | 33.0247 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 21.4859 | 17.2691 | oryzias_sinensis | ENSOSIP00000003303 | 45.9227 | 36.9099 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 36.9478 | 34.5382 | pelodiscus_sinensis | ENSPSIP00000002267 | 91.5423 | 85.5721 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 70.6827 | 64.6586 | gopherus_evgoodei | ENSGEVP00005025569 | 80.3653 | 73.516 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 35.9438 | 32.5301 | chelonoidis_abingdonii | ENSCABP00000015006 | 83.6449 | 75.7009 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 63.4538 | 52.8112 | seriola_dumerili | ENSSDUP00000027357 | 30.8895 | 25.7087 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 60.0402 | 49.3976 | cottoperca_gobio | ENSCGOP00000032544 | 29.3137 | 24.1176 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 45.3815 | 38.1526 | gasterosteus_aculeatus | ENSGACP00000001024 | 49.0239 | 41.2148 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 61.6466 | 49.5984 | cynoglossus_semilaevis | ENSCSEP00000027109 | 25.2675 | 20.3292 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 54.4177 | 44.5783 | poecilia_formosa | ENSPFOP00000002179 | 50.1852 | 41.1111 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 58.2329 | 48.7952 | myripristis_murdjan | ENSMMDP00005040823 | 31.4534 | 26.3557 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 62.249 | 54.2169 | strigops_habroptila | ENSSHBP00005008826 | 52.3649 | 45.6081 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 63.253 | 52.008 | amphiprion_ocellaris | ENSAOCP00000007742 | 30.2013 | 24.8322 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 30.5221 | 28.7149 | terrapene_carolina_triunguis | ENSTMTP00000026214 | 71.6981 | 67.4528 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 67.6707 | 61.245 | terrapene_carolina_triunguis | ENSTMTP00000010737 | 79.8578 | 72.2749 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 63.8554 | 54.2169 | hucho_hucho | ENSHHUP00000036639 | 48.6239 | 41.2844 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 32.7309 | 28.3133 | hucho_hucho | ENSHHUP00000010340 | 60.5948 | 52.4164 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 55.4217 | 45.5823 | betta_splendens | ENSBSLP00000023374 | 31.6877 | 26.062 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 62.6506 | 53.2129 | pygocentrus_nattereri | ENSPNAP00000018347 | 56.4195 | 47.9204 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 63.4538 | 51.4056 | anabas_testudineus | ENSATEP00000065567 | 31.4428 | 25.4726 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 8.03213 | 4.81928 | ciona_savignyi | ENSCSAVP00000001836 | 22.3464 | 13.4078 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 63.4538 | 52.8112 | seriola_lalandi_dorsalis | ENSSLDP00000007968 | 30.8895 | 25.7087 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 61.8474 | 50.8032 | labrus_bergylta | ENSLBEP00000016265 | 29.7297 | 24.4208 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 65.4618 | 53.2129 | pundamilia_nyererei | ENSPNYP00000027023 | 30.7838 | 25.0236 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 63.8554 | 52.008 | mastacembelus_armatus | ENSMAMP00000017412 | 30.8738 | 25.1456 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 63.0522 | 51.2048 | stegastes_partitus | ENSSPAP00000006876 | 32.2382 | 26.1807 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 60.241 | 51.8072 | oncorhynchus_tshawytscha | ENSOTSP00005012487 | 37.6412 | 32.3714 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 54.8193 | 46.988 | oncorhynchus_tshawytscha | ENSOTSP00005018157 | 50.369 | 43.1734 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 54.2169 | 45.7831 | acanthochromis_polyacanthus | ENSAPOP00000001343 | 50.7519 | 42.8571 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 37.1486 | 27.7108 | nothobranchius_furzeri | ENSNFUP00015047452 | 53.7791 | 40.1163 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 63.253 | 51.004 | nothobranchius_furzeri | ENSNFUP00015047459 | 28.4296 | 22.9242 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 55.0201 | 44.9799 | cyprinodon_variegatus | ENSCVAP00000020494 | 29.2423 | 23.9061 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 50.4016 | 39.759 | denticeps_clupeoides | ENSDCDP00000035452 | 51.3292 | 40.4908 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 62.6506 | 53.2129 | denticeps_clupeoides | ENSDCDP00000047314 | 56.6243 | 48.0944 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 57.2289 | 47.7912 | denticeps_clupeoides | ENSDCDP00000017465 | 59.874 | 50 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 62.8514 | 52.008 | larimichthys_crocea | ENSLCRP00005056075 | 30.4771 | 25.2191 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 61.0442 | 51.004 | takifugu_rubripes | ENSTRUP00000069482 | 30.7692 | 25.7085 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 60.241 | 48.1928 | hippocampus_comes | ENSHCOP00000012873 | 35.3774 | 28.3019 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 61.0442 | 51.4056 | cyprinus_carpio_carpio | ENSCCRP00000143551 | 55.1724 | 46.461 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 62.0482 | 52.4096 | cyprinus_carpio_carpio | ENSCCRP00000104751 | 56.5934 | 47.8022 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 62.249 | 51.004 | poecilia_latipinna | ENSPLAP00000026283 | 31.0933 | 25.4764 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 61.4458 | 52.2088 | sinocyclocheilus_grahami | ENSSGRP00000064983 | 56.6667 | 48.1481 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 61.6466 | 52.008 | sinocyclocheilus_grahami | ENSSGRP00000036940 | 56.7468 | 47.8743 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 65.4618 | 53.2129 | maylandia_zebra | ENSMZEP00005035839 | 30.7838 | 25.0236 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 44.7791 | 40.1606 | tetraodon_nigroviridis | ENSTNIP00000001935 | 51.6204 | 46.2963 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 67.2691 | 58.6345 | paramormyrops_kingsleyae | ENSPKIP00000008532 | 60.6884 | 52.8986 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 69.0763 | 59.4378 | scleropages_formosus | ENSSFOP00015067591 | 60.5634 | 52.1127 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 75.1004 | 66.2651 | leptobrachium_leishanense | ENSLLEP00000046749 | 81.3043 | 71.7391 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 63.0522 | 51.8072 | scophthalmus_maximus | ENSSMAP00000020301 | 29.4559 | 24.2026 |
| ENSG00000169057 | homo_sapiens | ENSP00000395535 | 60.0402 | 47.7912 | oryzias_melastigma | ENSOMEP00000009954 | 31.0811 | 24.7401 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0000122 | involved_in negative regulation of transcription by RNA polymerase II | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0000400 | enables four-way junction DNA binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0000792 | part_of heterochromatin | 2 | IBA,IDA | Homo_sapiens(9606) | Component |
| GO:0001662 | involved_in behavioral fear response | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0001666 | involved_in response to hypoxia | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0001964 | involved_in startle response | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0001976 | involved_in nervous system process involved in regulation of systemic arterial blood pressure | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0002087 | involved_in regulation of respiratory gaseous exchange by nervous system process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0003676 | enables nucleic acid binding | 1 | EXP | Homo_sapiens(9606) | Function |
| GO:0003677 | enables DNA binding | 1 | TAS | Homo_sapiens(9606) | Function |
| GO:0003682 | enables chromatin binding | 1 | IBA | Homo_sapiens(9606) | Function |
| GO:0003700 | enables DNA-binding transcription factor activity | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0003714 | enables transcription corepressor activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0003723 | enables RNA binding | 1 | HDA | Homo_sapiens(9606) | Function |
| GO:0003729 | enables mRNA binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005615 | located_in extracellular space | 1 | HDA | Homo_sapiens(9606) | Component |
| GO:0005634 | is_active_in nucleus | 2 | IBA,IDA | Homo_sapiens(9606) | Component |
| GO:0005654 | located_in nucleoplasm | 2 | IDA,TAS | Homo_sapiens(9606) | Component |
| GO:0005813 | located_in centrosome | 1 | IMP | Homo_sapiens(9606) | Component |
| GO:0005829 | located_in cytosol | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0006020 | involved_in inositol metabolic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0006349 | acts_upstream_of_or_within regulation of gene expression by genomic imprinting | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0006541 | involved_in glutamine metabolic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0006576 | involved_in biogenic amine metabolic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0007052 | involved_in mitotic spindle organization | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0007219 | involved_in Notch signaling pathway | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0007416 | involved_in synapse assembly | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0007507 | involved_in heart development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0007585 | involved_in respiratory gaseous exchange by respiratory system | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0007616 | involved_in long-term memory | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0008104 | involved_in protein localization | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0008211 | involved_in glucocorticoid metabolic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0008284 | involved_in positive regulation of cell population proliferation | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0008327 | enables methyl-CpG binding | 1 | IBA | Homo_sapiens(9606) | Function |
| GO:0008344 | involved_in adult locomotory behavior | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0008542 | involved_in visual learning | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0009791 | involved_in post-embryonic development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0010212 | involved_in response to ionizing radiation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0010288 | involved_in response to lead ion | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0010385 | enables double-stranded methylated DNA binding | 2 | IBA,IMP | Homo_sapiens(9606) | Function |
| GO:0010467 | involved_in gene expression | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0010629 | involved_in negative regulation of gene expression | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0010971 | involved_in positive regulation of G2/M transition of mitotic cell cycle | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0014003 | involved_in oligodendrocyte development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0014009 | involved_in glial cell proliferation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0016358 | involved_in dendrite development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0016525 | involved_in negative regulation of angiogenesis | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0016571 | involved_in histone methylation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0016573 | involved_in histone acetylation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0019230 | involved_in proprioception | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0019233 | involved_in sensory perception of pain | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0019904 | enables protein domain specific binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0021510 | involved_in spinal cord development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0021549 | involved_in cerebellum development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0021591 | involved_in ventricular system development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0021740 | involved_in principal sensory nucleus of trigeminal nerve development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0021756 | involved_in striatum development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0021766 | involved_in hippocampus development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0021772 | involved_in olfactory bulb development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0021794 | involved_in thalamus development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0021987 | involved_in cerebral cortex development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0031490 | enables chromatin DNA binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0031507 | involved_in heterochromatin formation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0031915 | involved_in positive regulation of synaptic plasticity | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0032048 | involved_in cardiolipin metabolic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0032091 | involved_in negative regulation of protein binding | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0032355 | involved_in response to estradiol | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0035176 | involved_in social behavior | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0035197 | enables siRNA binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0035865 | involved_in cellular response to potassium ion | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0042220 | involved_in response to cocaine | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0042551 | involved_in neuron maturation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0042826 | enables histone deacetylase binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0043524 | involved_in negative regulation of neuron apoptotic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0043537 | involved_in negative regulation of blood vessel endothelial cell migration | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0045322 | enables unmethylated CpG binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0045892 | involved_in negative regulation of DNA-templated transcription | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0045944 | involved_in positive regulation of transcription by RNA polymerase II | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0046470 | involved_in phosphatidylcholine metabolic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0048286 | involved_in lung alveolus development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0048712 | involved_in negative regulation of astrocyte differentiation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0050432 | involved_in catecholamine secretion | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0050807 | involved_in regulation of synapse organization | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0051151 | involved_in negative regulation of smooth muscle cell differentiation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0051707 | involved_in response to other organism | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0060079 | involved_in excitatory postsynaptic potential | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0060090 | enables molecular adaptor activity | 1 | EXP | Homo_sapiens(9606) | Function |
| GO:0060252 | involved_in positive regulation of glial cell proliferation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0060291 | involved_in long-term synaptic potentiation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0060999 | involved_in positive regulation of dendritic spine development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0061000 | involved_in negative regulation of dendritic spine development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0071317 | involved_in cellular response to isoquinoline alkaloid | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0090063 | involved_in positive regulation of microtubule nucleation | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0090327 | involved_in negative regulation of locomotion involved in locomotory behavior | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0098794 | located_in postsynapse | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0098978 | located_in glutamatergic synapse | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0099191 | involved_in trans-synaptic signaling by BDNF | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0099611 | involved_in regulation of action potential firing threshold | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0140693 | enables molecular condensate scaffold activity | 1 | IMP | Homo_sapiens(9606) | Function |
| GO:1900114 | involved_in positive regulation of histone H3-K9 trimethylation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:1901953 | involved_in positive regulation of anterograde dense core granule transport | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:1901956 | involved_in positive regulation of retrograde dense core granule transport | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:1903860 | involved_in negative regulation of dendrite extension | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:1903861 | involved_in positive regulation of dendrite extension | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:1903941 | involved_in negative regulation of respiratory gaseous exchange | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:1905492 | involved_in positive regulation of branching morphogenesis of a nerve | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:1905643 | involved_in positive regulation of DNA methylation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:1990841 | enables promoter-specific chromatin binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:2000635 | involved_in negative regulation of primary miRNA processing | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:2000820 | involved_in negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation | 1 | IEA | Homo_sapiens(9606) | Process |