Welcome to
RBP World!
Functions and Diseases
| RBP Type | Canonical_RBPs |
| Diseases | CancerFlavivirusesSARS-COV-2DengueZika |
| Drug | N.A. |
| Main interacting RNAs | mRNA |
| Moonlighting functions | N.A. |
| Localizations | N.A. |
| BulkPerturb-seq | N.A. |
Description
| Ensembl ID | ENSG00000162231 | Gene ID | 10482 | Accession | 8071 |
| Symbol | NXF1 | Alias | TAP;MEX67 | Full Name | nuclear RNA export factor 1 |
| Status | Confidence | Length | 14180 bases | Strand | Minus strand |
| Position | 11 : 62792123 - 62806302 | RNA binding domain | NTF2 , Tap-RNA_bind , TAP_C | ||
| Summary | This gene is one member of a family of nuclear RNA export factor genes. Common domain features of this family are a noncanonical RNP-type RNA-binding domain (RBD), 4 leucine-rich repeats (LRRs), a nuclear transport factor 2 (NTF2)-like domain that allows heterodimerization with NTF2-related export protein-1 (NXT1), and a ubiquitin-associated domain that mediates interactions with nucleoporins. The LRRs and NTF2-like domains are required for export activity. Alternative splicing seems to be a common mechanism in this gene family. The encoded protein of this gene shuttles between the nucleus and the cytoplasm and binds in vivo to poly(A)+ RNA. It is the vertebrate homologue of the yeast protein Mex67p. The encoded protein overcomes the mRNA export block caused by the presence of saturating amounts of CTE (constitutive transport element) RNA of type D retroviruses. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Jul 2008] | ||||
RNA binding domains (RBDs)
| Protein | Domain | Pfam ID | E-value | Domain number |
|---|---|---|---|---|
| ENSP00000453885 | Tap-RNA_bind | PF09162 | 9.7e-39 | 1 |
| ENSP00000435742 | Tap-RNA_bind | PF09162 | 1.8e-38 | 1 |
| ENSP00000435742 | NTF2 | PF02136 | 2.8e-27 | 2 |
| ENSP00000294172 | Tap-RNA_bind | PF09162 | 1.9e-38 | 1 |
| ENSP00000294172 | TAP_C | PF03943 | 4.3e-24 | 2 |
| ENSP00000294172 | NTF2 | PF02136 | 3.1e-27 | 3 |
| ENSP00000436679 | Tap-RNA_bind | PF09162 | 1.9e-38 | 1 |
| ENSP00000436679 | TAP_C | PF03943 | 4.3e-24 | 2 |
| ENSP00000436679 | NTF2 | PF02136 | 3.1e-27 | 3 |
| ENSP00000431381 | Tap-RNA_bind | PF09162 | 1.9e-29 | 1 |
| ENSP00000477375 | Tap-RNA_bind | PF09162 | 2.4e-24 | 1 |
| ENSP00000434301 | TAP_C | PF03943 | 3.7e-25 | 1 |
| ENSP00000434301 | NTF2 | PF02136 | 6.7e-06 | 2 |
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|---|---|---|---|
| 11356964 | Retroviral constitutive transport element evolved from cellular TAP(NXF1)-binding sequences. | S A Zolotukhin | 2001-06-01 | Journal of virology |
| 28977534 | Fractionation iCLIP detects persistent SR protein binding to conserved, retained introns in chromatin, nucleoplasm and cytoplasm. | Mattia Brugiolo | 2017-10-13 | Nucleic acids research |
| 27679854 | The role of TREX in gene expression and disease. | G Catherine Heath | 2016-10-01 | The Biochemical journal |
| 33033226 | Molecular mechanism underlying selective inhibition of mRNA nuclear export by herpesvirus protein ORF10. | Han Feng | 2020-10-27 | Proceedings of the National Academy of Sciences of the United States of America |
| 19834549 | Probing the HIV-1 genomic RNA trafficking pathway and dimerization by genetic recombination and single virion analyses. | D Michael Moore | 2009-10-01 | PLoS pathogens |
| 25605961 | Evolutionary conservation of a molecular machinery for export and expression of mRNAs with retained introns. | Baomin Wang | 2015-03-01 | RNA (New York, N.Y.) |
| 19019956 | Efficient nuclear export of herpes simplex virus 1 transcripts requires both RNA binding by ICP27 and ICP27 interaction with TAP/NXF1. | A Lisa Johnson | 2009-02-01 | Journal of virology |
| 18615016 | Drosophila RNAi screen identifies host genes important for influenza virus replication. | Linhui Hao | 2008-08-14 | Nature |
| 22505578 | Epstein-Barr virus protein EB2 stimulates cytoplasmic mRNA accumulation by counteracting the deleterious effects of SRp20 on viral mRNAs. | Franceline Juillard | 2012-08-01 | Nucleic acids research |
| 23951221 | A masked PY-NLS in Drosophila TIS11 and its mammalian homolog tristetraprolin. | Laure Twyffels | 2013-01-01 | PloS one |
| 24073193 | Highly pathogenic avian influenza virus nucleoprotein interacts with TREX complex adaptor protein Aly/REF. | T M R Vinod Balasubramaniam | 2013-01-01 | PloS one |
| 29508942 | Intron retention in viruses and cellular genes: Detention, border controls and passports. | David Rekosh | 2018-05-01 | Wiley interdisciplinary reviews. RNA |
| 15210956 | A molecular link between SR protein dephosphorylation and mRNA export. | Yingqun Huang | 2004-06-29 | Proceedings of the National Academy of Sciences of the United States of America |
| 20227104 | SR proteins SRp20 and 9G8 contribute to efficient export of herpes simplex virus 1 mRNAs. | Laurimar Escudero-Paunetto | 2010-06-05 | Virology |
| 33461637 | Nuclear mRNA maturation and mRNA export control: from trypanosomes to opisthokonts. | Susanne Kramer | 2021-09-01 | Parasitology |
| 31375530 | Imaging within single NPCs reveals NXF1's role in mRNA export on the cytoplasmic side of the pore. | Rakefet Ben-Yishay | 2019-09-02 | The Journal of cell biology |
| 34753509 | An ancient retroviral RNA element hidden in mammalian genomes and its involvement in co-opted retroviral gene regulation. | Koichi Kitao | 2021-11-10 | Retrovirology |
| 24550725 | Competitive and cooperative interactions mediate RNA transfer from herpesvirus saimiri ORF57 to the mammalian export adaptor ALYREF. | B Richard Tunnicliffe | 2014-02-01 | PLoS pathogens |
| 34793452 | Nucleoporin TPR promotes tRNA nuclear export and protein synthesis in lung cancer cells. | Miao Chen | 2021-11-01 | PLoS genetics |
| 36768958 | Non-Coding RNAs Regulating Mitochondrial Functions and the Oxidative Stress Response as Putative Targets against Age-Related Macular Degeneration (AMD). | T M Juha Hyttinen | 2023-01-30 | International journal of molecular sciences |
| 31940815 | The Ebola Virus Nucleoprotein Recruits the Nuclear RNA Export Factor NXF1 into Inclusion Bodies to Facilitate Viral Protein Expression. | Lisa Wendt | 2020-01-11 | Cells |
| 36593289 | De novo genes with an lncRNA origin encode unique human brain developmental functionality. | A Ni An | 2023-02-01 | Nature ecology & evolution |
| 29552131 | Integrated analysis of microarray data to identify the genes critical for the rupture of intracranial aneurysm. | Liang Wei | 2018-04-01 | Oncology letters |
| 30218090 | The m6A-methylase complex recruits TREX and regulates mRNA export. | Simon Lesbirel | 2018-09-14 | Scientific reports |
| 28592444 | Cellular differentiation state modulates the mRNA export activity of SR proteins. | Valentina Botti | 2017-07-03 | The Journal of cell biology |
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| NXF1-211 | ENST00000531709 | 4128 | 356aa | ENSP00000453885 |
| NXF1-201 | ENST00000294172 | 2290 | 619aa | ENSP00000294172 |
| NXF1-213 | ENST00000532297 | 2795 | 619aa | ENSP00000436679 |
| NXF1-206 | ENST00000527902 | 619 | 124aa | ENSP00000434301 |
| NXF1-214 | ENST00000533048 | 922 | No protein | - |
| NXF1-216 | ENST00000533499 | 936 | No protein | - |
| NXF1-205 | ENST00000527497 | 529 | No protein | - |
| NXF1-207 | ENST00000530875 | 1922 | 603aa | ENSP00000435742 |
| NXF1-215 | ENST00000533440 | 579 | No protein | - |
| NXF1-212 | ENST00000531872 | 390 | No protein | - |
| NXF1-210 | ENST00000531579 | 590 | No protein | - |
| NXF1-203 | ENST00000526163 | 569 | No protein | - |
| NXF1-208 | ENST00000531131 | 1023 | 219aa | ENSP00000477375 |
| NXF1-217 | ENST00000533671 | 589 | 115aa | ENSP00000431381 |
| NXF1-202 | ENST00000525576 | 572 | 76aa | ENSP00000432530 |
| NXF1-209 | ENST00000531474 | 548 | 63aa | ENSP00000437296 |
| NXF1-204 | ENST00000527064 | 572 | No protein | - |
Phenotypes
| ensgID | Trait | pValue | Pubmed ID |
|---|
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
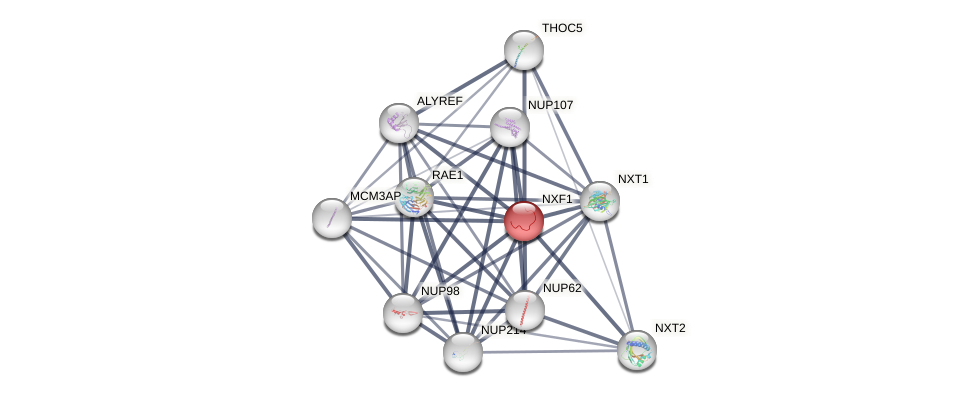
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000162231 | homo_sapiens | ENSP00000472530 | 72.0447 | 57.508 | homo_sapiens | ENSP00000294172 | 72.8595 | 58.1583 |
| ENSG00000162231 | homo_sapiens | ENSP00000378504 | 66.4783 | 50.8475 | homo_sapiens | ENSP00000294172 | 57.0275 | 43.6187 |
| ENSG00000162231 | homo_sapiens | ENSP00000485586 | 72.0447 | 57.508 | homo_sapiens | ENSP00000294172 | 72.8595 | 58.1583 |
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 65.105 | 65.105 | nomascus_leucogenys | ENSNLEP00000005047 | 99.7525 | 99.7525 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 100 | 100 | pan_paniscus | ENSPPAP00000037493 | 100 | 100 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 99.5154 | 99.3538 | pongo_abelii | ENSPPYP00000003631 | 99.5154 | 99.3538 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 100 | 100 | pan_troglodytes | ENSPTRP00000006511 | 100 | 100 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 100 | 99.8384 | gorilla_gorilla | ENSGGOP00000017691 | 100 | 99.8384 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 89.1761 | 80.2908 | ornithorhynchus_anatinus | ENSOANP00000041596 | 71.134 | 64.0464 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 91.5994 | 83.1987 | sarcophilus_harrisii | ENSSHAP00000007371 | 88.4555 | 80.3432 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 80.937 | 66.0743 | sarcophilus_harrisii | ENSSHAP00000001998 | 76.6055 | 62.5382 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 75.1212 | 68.1745 | notamacropus_eugenii | ENSMEUP00000012106 | 74.8792 | 67.9549 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 50.2423 | 40.8724 | notamacropus_eugenii | ENSMEUP00000004344 | 87.8531 | 71.4689 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 96.4459 | 92.5687 | dasypus_novemcinctus | ENSDNOP00000017447 | 96.6019 | 92.7184 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 96.9305 | 92.4071 | erinaceus_europaeus | ENSEEUP00000002181 | 97.0874 | 92.5566 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 75.7674 | 70.1131 | echinops_telfairi | ENSETEP00000009178 | 77.3927 | 71.6172 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 99.3538 | 98.3845 | callithrix_jacchus | ENSCJAP00000093824 | 99.3538 | 98.3845 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 100 | 100 | cercocebus_atys | ENSCATP00000042951 | 100 | 100 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 100 | 100 | macaca_fascicularis | ENSMFAP00000028107 | 100 | 100 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 100 | 100 | macaca_mulatta | ENSMMUP00000000265 | 100 | 100 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 100 | 100 | macaca_nemestrina | ENSMNEP00000021941 | 100 | 100 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 100 | 100 | papio_anubis | ENSPANP00000011419 | 100 | 100 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 100 | 100 | mandrillus_leucophaeus | ENSMLEP00000006490 | 100 | 100 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 87.5606 | 84.0065 | tursiops_truncatus | ENSTTRP00000002326 | 87.7023 | 84.1424 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 96.1228 | 92.5687 | vulpes_vulpes | ENSVVUP00000041987 | 84.517 | 81.392 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 97.0921 | 93.6995 | equus_asinus | ENSEASP00005016417 | 97.2492 | 93.8511 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 97.0921 | 92.5687 | capra_hircus | ENSCHIP00000006269 | 93.0341 | 88.6997 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 96.1228 | 93.0533 | loxodonta_africana | ENSLAFP00000005699 | 96.4344 | 93.3549 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 97.8998 | 94.9919 | panthera_leo | ENSPLOP00000010995 | 98.0583 | 95.1456 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 98.0614 | 94.1842 | ailuropoda_melanoleuca | ENSAMEP00000035370 | 91.1411 | 87.5375 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 97.2536 | 93.0533 | bos_indicus_hybrid | ENSBIXP00005005778 | 97.0968 | 92.9032 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 95.4766 | 91.5994 | oryctolagus_cuniculus | ENSOCUP00000036082 | 89.5455 | 85.9091 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 88.0452 | 85.622 | equus_caballus | ENSECAP00000003616 | 97.4955 | 94.8122 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 96.9305 | 93.3764 | canis_lupus_familiaris | ENSCAFP00845010817 | 95.0872 | 91.6006 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 97.8998 | 94.9919 | panthera_pardus | ENSPPRP00000010065 | 98.0583 | 95.1456 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 97.0921 | 92.8918 | marmota_marmota_marmota | ENSMMMP00000003785 | 97.0921 | 92.8918 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 96.1228 | 92.4071 | balaenoptera_musculus | ENSBMSP00010011816 | 90.9786 | 87.4618 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 98.3845 | 95.4766 | cavia_porcellus | ENSCPOP00000005526 | 98.5437 | 95.6311 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 97.8998 | 94.9919 | felis_catus | ENSFCAP00000013705 | 98.0583 | 95.1456 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 93.3764 | 89.0145 | ursus_americanus | ENSUAMP00000034074 | 93.2258 | 88.871 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 91.7609 | 83.1987 | monodelphis_domestica | ENSMODP00000010059 | 91.0256 | 82.5321 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 78.0291 | 60.5816 | monodelphis_domestica | ENSMODP00000027677 | 73.1818 | 56.8182 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 95.6381 | 91.5994 | bison_bison_bison | ENSBBBP00000011688 | 95.4839 | 91.4516 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 96.1228 | 92.7302 | monodon_monoceros | ENSMMNP00015027304 | 95.9677 | 92.5806 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 95.6381 | 91.7609 | sus_scrofa | ENSSSCP00000060958 | 82.1082 | 78.7795 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 99.0307 | 97.4152 | microcebus_murinus | ENSMICP00000038752 | 99.1909 | 97.5728 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 96.2843 | 90.6301 | octodon_degus | ENSODEP00000002072 | 98.0263 | 92.2697 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 95.9612 | 92.7302 | jaculus_jaculus | ENSJJAP00000015280 | 95.4984 | 92.283 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 97.0921 | 92.7302 | ovis_aries_rambouillet | ENSOARP00020027434 | 96.9355 | 92.5806 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 97.0921 | 92.8918 | ursus_maritimus | ENSUMAP00000019876 | 94.795 | 90.694 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 72.2132 | 69.9515 | ochotona_princeps | ENSOPRP00000002171 | 72.3301 | 70.0647 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 96.1228 | 92.4071 | delphinapterus_leucas | ENSDLEP00000025101 | 87.6289 | 84.2415 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 95.315 | 92.2456 | physeter_catodon | ENSPCTP00005024172 | 96.7213 | 93.6066 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 95.1535 | 90.6301 | mus_caroli | MGP_CAROLIEiJ_P0048013 | 95.3074 | 90.7767 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 95.1535 | 91.1147 | mus_musculus | ENSMUSP00000010241 | 95.3074 | 91.2621 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 76.5751 | 73.8288 | dipodomys_ordii | ENSDORP00000025734 | 89.434 | 86.2264 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 97.5767 | 93.8611 | mustela_putorius_furo | ENSMPUP00000013555 | 97.7346 | 94.0129 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 98.0614 | 96.2843 | aotus_nancymaae | ENSANAP00000013420 | 97.2756 | 95.5128 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 99.1922 | 98.2229 | saimiri_boliviensis_boliviensis | ENSSBOP00000005609 | 99.1922 | 98.2229 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 100 | 99.8384 | chlorocebus_sabaeus | ENSCSAP00000003024 | 100 | 99.8384 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 77.0598 | 75.4443 | tupaia_belangeri | ENSTBEP00000011841 | 81.6781 | 79.9658 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 96.6074 | 94.1842 | mesocricetus_auratus | ENSMAUP00000014994 | 96.7638 | 94.3366 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 95.7997 | 91.7609 | phocoena_sinus | ENSPSNP00000026902 | 95.6452 | 91.6129 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 97.0921 | 93.6995 | camelus_dromedarius | ENSCDRP00005006949 | 97.2492 | 93.8511 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 98.3845 | 96.769 | carlito_syrichta | ENSTSYP00000019260 | 98.5437 | 96.9256 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 96.1228 | 93.2149 | panthera_tigris_altaica | ENSPTIP00000007248 | 96.1228 | 93.2149 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 94.5073 | 91.4378 | rattus_norvegicus | ENSRNOP00000081812 | 94.5073 | 91.4378 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 98.7076 | 97.2536 | prolemur_simus | ENSPSMP00000033482 | 98.8673 | 97.411 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 94.9919 | 90.4685 | microtus_ochrogaster | ENSMOCP00000001249 | 95.4545 | 90.9091 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 68.4976 | 66.559 | sorex_araneus | ENSSARP00000012489 | 85.4839 | 83.0645 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 94.1842 | 89.8223 | peromyscus_maniculatus_bairdii | ENSPEMP00000004124 | 94.4895 | 90.1134 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 97.2536 | 93.0533 | bos_mutus | ENSBMUP00000031531 | 97.0968 | 92.9032 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 95.315 | 91.2763 | mus_spicilegus | ENSMSIP00000037457 | 95.4693 | 91.424 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 96.769 | 93.8611 | cricetulus_griseus_chok1gshd | ENSCGRP00001012297 | 96.9256 | 94.0129 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 97.0921 | 93.8611 | pteropus_vampyrus | ENSPVAP00000003306 | 97.2492 | 94.0129 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 95.7997 | 91.7609 | myotis_lucifugus | ENSMLUP00000010164 | 95.7997 | 91.7609 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 92.5687 | 83.5218 | vombatus_ursinus | ENSVURP00010001723 | 91.8269 | 82.8526 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 93.538 | 89.3376 | rhinolophus_ferrumequinum | ENSRFEP00010017759 | 93.0868 | 88.9068 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 96.769 | 93.0533 | neovison_vison | ENSNVIP00000026643 | 94.4795 | 90.8517 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 92.4071 | 87.8837 | bos_taurus | ENSBTAP00000012486 | 91.3738 | 86.901 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 95.4766 | 91.9225 | heterocephalus_glaber_female | ENSHGLP00000003989 | 96.0976 | 92.5203 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 100 | 100 | rhinopithecus_bieti | ENSRBIP00000037915 | 100 | 100 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 97.7383 | 94.5073 | canis_lupus_dingo | ENSCAFP00020016383 | 97.8964 | 94.6602 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 97.4152 | 94.8304 | sciurus_vulgaris | ENSSVLP00005006121 | 97.5728 | 94.9838 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 49.7577 | 36.5105 | phascolarctos_cinereus | ENSPCIP00000042550 | 61.7234 | 45.2906 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 92.7302 | 84.3296 | phascolarctos_cinereus | ENSPCIP00000019977 | 91.9872 | 83.6538 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 50.0808 | 39.58 | phascolarctos_cinereus | ENSPCIP00000050180 | 78.2828 | 61.8687 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 94.8304 | 91.1147 | procavia_capensis | ENSPCAP00000015554 | 95.1378 | 91.41 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 96.1228 | 94.1842 | nannospalax_galili | ENSNGAP00000002130 | 96.1228 | 94.1842 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 98.3845 | 95.1535 | chinchilla_lanigera | ENSCLAP00000002050 | 98.5437 | 95.3074 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 95.6381 | 91.7609 | cervus_hanglu_yarkandensis | ENSCHYP00000028078 | 95.0241 | 91.1718 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 96.769 | 92.7302 | catagonus_wagneri | ENSCWAP00000024155 | 77.9948 | 74.7396 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 94.1842 | 90.1454 | ictidomys_tridecemlineatus | ENSSTOP00000027740 | 93.131 | 89.1374 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 96.1228 | 93.6995 | otolemur_garnettii | ENSOGAP00000007983 | 95.9677 | 93.5484 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 99.3538 | 98.3845 | cebus_imitator | ENSCCAP00000025617 | 99.3538 | 98.3845 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 95.9612 | 92.2456 | urocitellus_parryii | ENSUPAP00010006678 | 62.395 | 59.979 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 49.7577 | 45.7189 | moschus_moschiferus | ENSMMSP00000004373 | 87.0057 | 79.9435 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 96.2843 | 91.2763 | moschus_moschiferus | ENSMMSP00000016353 | 93.4169 | 88.558 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 100 | 100 | rhinopithecus_roxellana | ENSRROP00000041969 | 100 | 100 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 95.6381 | 92.2456 | mus_pahari | MGP_PahariEiJ_P0021017 | 95.9481 | 92.5446 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 98.7076 | 97.0921 | propithecus_coquereli | ENSPCOP00000024507 | 98.8673 | 97.2492 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 32.7948 | 18.0937 | caenorhabditis_elegans | C15H11.6.1 | 50.1235 | 27.6543 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 42.9725 | 24.8788 | caenorhabditis_elegans | C15H11.3.1 | 43.6782 | 25.2874 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 39.58 | 21.6478 | drosophila_melanogaster | FBpp0424323 | 39.8374 | 21.7886 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 21.9709 | 13.2472 | drosophila_melanogaster | FBpp0081102 | 45.1827 | 27.2425 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 51.6963 | 32.1486 | drosophila_melanogaster | FBpp0073267 | 47.619 | 29.6131 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 43.6187 | 24.7173 | drosophila_melanogaster | FBpp0088483 | 32.1046 | 18.1926 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 52.8271 | 33.1179 | ciona_intestinalis | ENSCINP00000026482 | 53.6066 | 33.6066 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 29.4023 | 24.7173 | petromyzon_marinus | ENSPMAP00000009507 | 71.9368 | 60.4743 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 51.5347 | 39.4184 | eptatretus_burgeri | ENSEBUP00000022462 | 76.4988 | 58.5132 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 15.1858 | 10.8239 | callorhinchus_milii | ENSCMIP00000004892 | 55.9524 | 39.881 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 86.5913 | 72.3748 | latimeria_chalumnae | ENSLACP00000001374 | 85.3503 | 71.3376 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 65.4281 | 52.3425 | lepisosteus_oculatus | ENSLOCP00000001429 | 78.4884 | 62.7907 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 73.5057 | 61.5509 | clupea_harengus | ENSCHAP00000022511 | 72.6837 | 60.8626 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 73.9903 | 59.1276 | clupea_harengus | ENSCHAP00000059361 | 71.118 | 56.8323 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 66.0743 | 48.7884 | danio_rerio | ENSDARP00000109457 | 67.0492 | 49.5082 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 78.3522 | 63.0048 | danio_rerio | ENSDARP00000071848 | 75.5452 | 60.7477 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 70.7593 | 53.6349 | carassius_auratus | ENSCARP00000061706 | 67.6971 | 51.3138 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 73.021 | 58.4814 | carassius_auratus | ENSCARP00000132498 | 69.7531 | 55.8642 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 73.8288 | 58.9661 | carassius_auratus | ENSCARP00000132695 | 71.2949 | 56.9423 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 72.5363 | 58.1583 | carassius_auratus | ENSCARP00000067644 | 69.8289 | 55.9876 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 73.9903 | 60.5816 | astyanax_mexicanus | ENSAMXP00000047207 | 71.3396 | 58.4112 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 76.0905 | 62.6817 | astyanax_mexicanus | ENSAMXP00000051323 | 71.148 | 58.6103 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 71.2439 | 56.3813 | ictalurus_punctatus | ENSIPUP00000029523 | 67.0213 | 53.0395 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 75.4443 | 60.9047 | ictalurus_punctatus | ENSIPUP00000030918 | 73.6593 | 59.4637 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 76.7367 | 62.1971 | esox_lucius | ENSELUP00000070353 | 75.8786 | 61.5016 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 61.7124 | 48.1422 | esox_lucius | ENSELUP00000059931 | 72.7619 | 56.7619 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 74.9596 | 61.0662 | electrophorus_electricus | ENSEEEP00000034455 | 73.5341 | 59.9049 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 71.8901 | 58.9661 | electrophorus_electricus | ENSEEEP00000039630 | 69.6401 | 57.1205 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 76.4136 | 61.7124 | oncorhynchus_kisutch | ENSOKIP00005089706 | 74.7235 | 60.3475 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 64.6204 | 51.5347 | oncorhynchus_kisutch | ENSOKIP00005030542 | 75.9013 | 60.5313 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 76.4136 | 61.2278 | oncorhynchus_kisutch | ENSOKIP00005092984 | 74.4882 | 59.685 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 75.1212 | 60.42 | oncorhynchus_mykiss | ENSOMYP00000020081 | 74.2812 | 59.7444 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 76.4136 | 61.5509 | oncorhynchus_mykiss | ENSOMYP00000072162 | 74.6057 | 60.0946 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 77.8675 | 62.5202 | oncorhynchus_mykiss | ENSOMYP00000096452 | 76.3867 | 61.3312 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 75.6058 | 61.3893 | salmo_salar | ENSSSAP00000089923 | 72.4458 | 58.8235 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 74.9596 | 59.6123 | salmo_salar | ENSSSAP00000051811 | 70.948 | 56.422 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 77.8675 | 62.6817 | salmo_salar | ENSSSAP00000051513 | 76.2658 | 61.3924 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 76.5751 | 61.5509 | salmo_trutta | ENSSTUP00000074148 | 73.7169 | 59.2535 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 75.9289 | 61.2278 | salmo_trutta | ENSSTUP00000057267 | 74.0157 | 59.685 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 69.79 | 55.7351 | salmo_trutta | ENSSTUP00000002820 | 75.5245 | 60.3147 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 76.4136 | 60.2585 | gadus_morhua | ENSGMOP00000029329 | 68.9504 | 54.3732 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 76.7367 | 58.3199 | fundulus_heteroclitus | ENSFHEP00000022187 | 72.8528 | 55.3681 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 77.706 | 59.7738 | poecilia_reticulata | ENSPREP00000022675 | 72.1139 | 55.4723 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 74.6365 | 58.3199 | xiphophorus_maculatus | ENSXMAP00000003174 | 73.2171 | 57.2108 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 77.2213 | 59.2892 | oryzias_latipes | ENSORLP00000003062 | 71.8797 | 55.188 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 75.9289 | 59.6123 | cyclopterus_lumpus | ENSCLMP00005018923 | 74.2496 | 58.2938 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 78.5137 | 61.2278 | oreochromis_niloticus | ENSONIP00000035779 | 73.7481 | 57.5114 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 78.0291 | 61.5509 | haplochromis_burtoni | ENSHBUP00000011267 | 75.5869 | 59.6244 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 77.3829 | 61.3893 | astatotilapia_calliptera | ENSACLP00000025270 | 73.2416 | 58.104 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 71.7286 | 56.5428 | sparus_aurata | ENSSAUP00010045339 | 74.6218 | 58.8235 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 77.706 | 61.0662 | lates_calcarifer | ENSLCAP00010043462 | 67.9379 | 53.3898 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 75.2827 | 60.0969 | xenopus_tropicalis | ENSXETP00000087233 | 75.0403 | 59.9034 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 86.5913 | 72.8595 | chrysemys_picta_bellii | ENSCPBP00000034135 | 84.9445 | 71.4739 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 84.168 | 70.4362 | sphenodon_punctatus | ENSSPUP00000010374 | 82.6984 | 69.2064 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 71.0824 | 55.7351 | laticauda_laticaudata | ENSLLTP00000002110 | 73.5786 | 57.6923 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 75.9289 | 61.874 | notechis_scutatus | ENSNSUP00000017690 | 77.9436 | 63.5158 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 67.8514 | 53.3118 | pseudonaja_textilis | ENSPTXP00000013574 | 77.4908 | 60.8856 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 73.5057 | 64.2973 | anas_platyrhynchos_platyrhynchos | ENSAPLP00000020998 | 69.4657 | 60.7634 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 85.7835 | 75.6058 | gallus_gallus | ENSGALP00010001536 | 71.8539 | 63.3288 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 70.2746 | 58.3199 | anolis_carolinensis | ENSACAP00000015638 | 71.1948 | 59.0835 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 78.5137 | 62.3586 | neolamprologus_brichardi | ENSNBRP00000001461 | 75.2322 | 59.7523 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 79.9677 | 65.7512 | naja_naja | ENSNNAP00000013714 | 65.7371 | 54.0505 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 75.4443 | 63.0048 | podarcis_muralis | ENSPMRP00000027489 | 79.6928 | 66.5529 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 84.168 | 71.8901 | taeniopygia_guttata | ENSTGUP00000019280 | 45.5818 | 38.9326 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 81.0985 | 69.1438 | taeniopygia_guttata | ENSTGUP00000029743 | 51.6993 | 44.0783 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 77.2213 | 64.7819 | erpetoichthys_calabaricus | ENSECRP00000015602 | 74.8044 | 62.7543 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 77.706 | 60.9047 | kryptolebias_marmoratus | ENSKMAP00000005355 | 72.549 | 56.8627 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 79.3215 | 63.0048 | salvator_merianae | ENSSMRP00000013356 | 78.56 | 62.4 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 76.252 | 58.8045 | oryzias_javanicus | ENSOJAP00000027969 | 70.5531 | 54.4096 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 77.706 | 61.5509 | amphilophus_citrinellus | ENSACIP00000029118 | 76.2282 | 60.3803 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 66.8821 | 52.3425 | dicentrarchus_labrax | ENSDLAP00005009715 | 68.0921 | 53.2895 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 76.5751 | 59.9354 | amphiprion_percula | ENSAPEP00000013771 | 74.7634 | 58.5173 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 77.2213 | 59.2892 | oryzias_sinensis | ENSOSIP00000043810 | 71.7718 | 55.1051 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 81.7448 | 68.6591 | pelodiscus_sinensis | ENSPSIP00000006879 | 82.9508 | 69.6721 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 85.4604 | 72.2132 | gopherus_evgoodei | ENSGEVP00005011485 | 70.6275 | 59.6796 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 85.2989 | 71.4055 | chelonoidis_abingdonii | ENSCABP00000029721 | 83.5443 | 69.9367 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 77.706 | 61.2278 | seriola_dumerili | ENSSDUP00000023805 | 71.791 | 56.5672 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 74.9596 | 58.3199 | cottoperca_gobio | ENSCGOP00000028810 | 73.4177 | 57.1203 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 86.9144 | 77.2213 | anser_brachyrhynchus | ENSABRP00000025453 | 86.7742 | 77.0968 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 76.5751 | 59.9354 | gasterosteus_aculeatus | ENSGACP00000022397 | 75 | 58.7025 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 77.8675 | 61.5509 | cynoglossus_semilaevis | ENSCSEP00000009015 | 74.0399 | 58.5253 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 77.8675 | 60.0969 | poecilia_formosa | ENSPFOP00000009319 | 77.12 | 59.52 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 67.5283 | 53.6349 | myripristis_murdjan | ENSMMDP00005043985 | 72.4437 | 57.539 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 78.5137 | 61.0662 | sander_lucioperca | ENSSLUP00000038222 | 72.6457 | 56.5022 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 85.622 | 74.475 | strigops_habroptila | ENSSHBP00005009078 | 83.7283 | 72.8278 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 75.1212 | 58.4814 | amphiprion_ocellaris | ENSAOCP00000012462 | 75.487 | 58.7662 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 86.7528 | 73.3441 | terrapene_carolina_triunguis | ENSTMTP00000009278 | 84.8341 | 71.722 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 76.5751 | 61.874 | hucho_hucho | ENSHHUP00000082710 | 74.8815 | 60.5055 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 75.1212 | 60.2585 | hucho_hucho | ENSHHUP00000021025 | 70.4545 | 56.5152 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 77.706 | 62.3586 | hucho_hucho | ENSHHUP00000041387 | 75.9874 | 60.9795 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 77.3829 | 60.7431 | betta_splendens | ENSBSLP00000042649 | 73.5791 | 57.7573 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 76.4136 | 62.1971 | pygocentrus_nattereri | ENSPNAP00000004701 | 74.3711 | 60.5346 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 75.7674 | 61.2278 | pygocentrus_nattereri | ENSPNAP00000020114 | 73.9748 | 59.7792 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 77.8675 | 60.42 | anabas_testudineus | ENSATEP00000047535 | 74.4977 | 57.8053 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 46.042 | 28.5945 | ciona_savignyi | ENSCSAVP00000013296 | 58.0448 | 36.0489 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 41.1955 | 22.294 | saccharomyces_cerevisiae | YPL169C | 42.571 | 23.0384 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 72.2132 | 56.8659 | seriola_lalandi_dorsalis | ENSSLDP00000020559 | 79.6791 | 62.7451 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 78.0291 | 60.7431 | labrus_bergylta | ENSLBEP00000007770 | 75.1166 | 58.4759 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 78.5137 | 62.1971 | pundamilia_nyererei | ENSPNYP00000019444 | 77.1429 | 61.1111 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 74.7981 | 59.1276 | mastacembelus_armatus | ENSMAMP00000055403 | 73.3756 | 58.0032 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 68.6591 | 52.6656 | stegastes_partitus | ENSSPAP00000009625 | 72.8988 | 55.9177 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 76.4136 | 61.7124 | oncorhynchus_tshawytscha | ENSOTSP00005068160 | 74.6057 | 60.2524 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 77.8675 | 62.5202 | oncorhynchus_tshawytscha | ENSOTSP00005019067 | 76.3867 | 61.3312 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 73.6672 | 58.4814 | oncorhynchus_tshawytscha | ENSOTSP00005079091 | 69.3009 | 55.0152 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 76.7367 | 60.0969 | acanthochromis_polyacanthus | ENSAPOP00000013908 | 75.0395 | 58.7678 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 86.5913 | 76.252 | aquila_chrysaetos_chrysaetos | ENSACCP00020017859 | 87.1545 | 76.748 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 75.7674 | 59.9354 | nothobranchius_furzeri | ENSNFUP00015012515 | 75.04 | 59.36 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 75.2827 | 58.8045 | cyprinodon_variegatus | ENSCVAP00000005068 | 75.1613 | 58.7097 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 73.5057 | 59.4507 | denticeps_clupeoides | ENSDCDP00000016821 | 73.7439 | 59.6434 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 73.5057 | 59.2892 | denticeps_clupeoides | ENSDCDP00000017320 | 73.9837 | 59.6748 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 75.7674 | 61.3893 | denticeps_clupeoides | ENSDCDP00000042528 | 75.04 | 60.8 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 78.1906 | 60.9047 | larimichthys_crocea | ENSLCRP00005051955 | 73.0015 | 56.8627 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 76.4136 | 59.7738 | takifugu_rubripes | ENSTRUP00000077902 | 75.8013 | 59.2949 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 64.1357 | 49.4346 | hippocampus_comes | ENSHCOP00000028196 | 77.6908 | 59.8826 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 33.1179 | 22.294 | cyprinus_carpio_carpio | ENSCCRP00000142842 | 55.4054 | 37.2973 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 74.9596 | 60.9047 | cyprinus_carpio_carpio | ENSCCRP00000131326 | 73.5341 | 59.7464 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 70.1131 | 53.1502 | cyprinus_carpio_carpio | ENSCCRP00000041152 | 69.1083 | 52.3885 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 76.4136 | 62.3586 | cyprinus_carpio_carpio | ENSCCRP00000172306 | 70.2823 | 57.3551 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 78.0291 | 60.0969 | poecilia_latipinna | ENSPLAP00000031456 | 72.1973 | 55.6054 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 49.273 | 37.8029 | sinocyclocheilus_grahami | ENSSGRP00000072357 | 63.8075 | 48.954 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 77.8675 | 63.3279 | sinocyclocheilus_grahami | ENSSGRP00000106359 | 75.6672 | 61.5385 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 77.5444 | 63.3279 | sinocyclocheilus_grahami | ENSSGRP00000069622 | 74.1886 | 60.5873 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 46.2036 | 33.1179 | sinocyclocheilus_grahami | ENSSGRP00000039670 | 56.2992 | 40.3543 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 77.3829 | 61.3893 | maylandia_zebra | ENSMZEP00005006352 | 73.2416 | 58.104 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 56.0582 | 42.6494 | tetraodon_nigroviridis | ENSTNIP00000019915 | 70.101 | 53.3333 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 15.5089 | 12.9241 | tetraodon_nigroviridis | ENSTNIP00000003145 | 82.0513 | 68.3761 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 80.2908 | 65.5897 | paramormyrops_kingsleyae | ENSPKIP00000010537 | 79.1401 | 64.6497 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 79.9677 | 66.3974 | scleropages_formosus | ENSSFOP00015011633 | 77.9528 | 64.7244 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 65.5897 | 50.2423 | leptobrachium_leishanense | ENSLLEP00000044236 | 68.4654 | 52.4452 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 27.1405 | 19.5477 | leptobrachium_leishanense | ENSLLEP00000044413 | 60.8696 | 43.8406 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 72.0517 | 55.8966 | leptobrachium_leishanense | ENSLLEP00000040662 | 70.4581 | 54.6603 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 64.9435 | 50.0808 | leptobrachium_leishanense | ENSLLEP00000044516 | 66.3366 | 51.1551 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 75.2827 | 59.7738 | scophthalmus_maximus | ENSSMAP00000050587 | 69.6562 | 55.3064 |
| ENSG00000162231 | homo_sapiens | ENSP00000294172 | 77.2213 | 59.2892 | oryzias_melastigma | ENSOMEP00000016827 | 71.988 | 55.2711 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0003723 | enables RNA binding | 2 | HDA,IBA | Homo_sapiens(9606) | Function |
| GO:0003729 | enables mRNA binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005634 | is_active_in nucleus | 3 | IBA,IDA,ISS | Homo_sapiens(9606) | Component |
| GO:0005643 | part_of nuclear pore | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0005654 | located_in nucleoplasm | 2 | IDA,TAS | Homo_sapiens(9606) | Component |
| GO:0005737 | located_in cytoplasm | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005829 | located_in cytosol | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0006406 | involved_in mRNA export from nucleus | 2 | IDA,ISS | Homo_sapiens(9606) | Process |
| GO:0010494 | located_in cytoplasmic stress granule | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0015031 | involved_in protein transport | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0016607 | located_in nuclear speck | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0016973 | involved_in poly(A)+ mRNA export from nucleus | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0042272 | part_of nuclear RNA export factor complex | 1 | IPI | Homo_sapiens(9606) | Component |
| GO:0042405 | located_in nuclear inclusion body | 1 | IEA | Homo_sapiens(9606) | Component |