Welcome to
RBP World!
Functions and Diseases
| RBP Type | Non-canonical_RBPs |
| Diseases | CancerSARS-COV-2 |
| Drug | N.A. |
| Main interacting RNAs | N.A. |
| Moonlighting functions | E3 ligase |
| Localizations | Stress granucle |
| BulkPerturb-seq | N.A. |
Description
| Ensembl ID | ENSG00000161011 | Gene ID | 8878 | Accession | 11280 |
| Symbol | SQSTM1 | Alias | p60;p62;A170;DMRV;OSIL;PDB3;ZIP3;p62B;NADGP;FTDALS3 | Full Name | sequestosome 1 |
| Status | Confidence | Length | 31671 bases | Strand | Plus strand |
| Position | 5 : 179806408 - 179838078 | RNA binding domain | N.A. | ||
| Summary | This gene encodes a multifunctional protein that binds ubiquitin and regulates activation of the nuclear factor kappa-B (NF-kB) signaling pathway. The protein functions as a scaffolding/adaptor protein in concert with TNF receptor-associated factor 6 to mediate activation of NF-kB in response to upstream signals. Alternatively spliced transcript variants encoding either the same or different isoforms have been identified for this gene. Mutations in this gene result in sporadic and familial Paget disease of bone. [provided by RefSeq, Mar 2009] | ||||
RNA binding domains (RBDs)
| Protein | Domain | Pfam ID | E-value | Domain number |
|---|
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|---|---|---|---|
| 31006338 | Vault RNA emerges as a regulator of selective autophagy. | Rastislav Horos | 2019-08-01 | Autophagy |
| 31097692 | Impaired autophagic degradation of lncRNA ARHGAP5-AS1 promotes chemoresistance in gastric cancer. | Liyuan Zhu | 2019-05-16 | Cell death & disease |
| 33662035 | The CDT of Helicobacter hepaticus induces pro-survival autophagy and nucleoplasmic reticulum formation concentrating the RNA binding proteins UNR/CSDE1 and P62/SQSTM1. | Wencan He | 2021-03-01 | PLoS pathogens |
| 31753861 | Autophagy and cancer: Modulation of cell death pathways and cancer cell adaptations. | G Christina Towers | 2020-01-06 | The Journal of cell biology |
| 22174682 | IRGM is a common target of RNA viruses that subvert the autophagy network. | Pombo Isabel Grégoire | 2011-12-01 | PLoS pathogens |
| 23922739 | Autophagy activation clears ELAVL1/HuR-mediated accumulation of SQSTM1/p62 during proteasomal inhibition in human retinal pigment epithelial cells. | Johanna Viiri | 2013-01-01 | PloS one |
| 30979586 | ULK1 and ULK2 Regulate Stress Granule Disassembly Through Phosphorylation and Activation of VCP/p97. | Bo Wang | 2019-05-16 | Molecular cell |
| 35545049 | The lactate-NAD+ axis activates cancer-associated fibroblasts by downregulating p62. | F Juan Linares | 2022-05-10 | Cell reports |
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| SQSTM1-210 | ENST00000506042 | 921 | No protein | - |
| SQSTM1-215 | ENST00000514093 | 842 | 167aa | ENSP00000427308 |
| SQSTM1-211 | ENST00000506690 | 2405 | No protein | - |
| SQSTM1-214 | ENST00000512234 | 1722 | No protein | - |
| SQSTM1-203 | ENST00000422245 | 628 | 140aa | ENSP00000394534 |
| SQSTM1-207 | ENST00000481335 | 562 | No protein | - |
| SQSTM1-205 | ENST00000464493 | 532 | No protein | - |
| SQSTM1-212 | ENST00000508284 | 578 | 69aa | ENSP00000424195 |
| SQSTM1-216 | ENST00000626660 | 274 | 73aa | ENSP00000487071 |
| SQSTM1-204 | ENST00000453046 | 572 | 73aa | ENSP00000405061 |
| SQSTM1-209 | ENST00000504627 | 602 | 190aa | ENSP00000425957 |
| SQSTM1-202 | ENST00000389805 | 2840 | 440aa | ENSP00000374455 |
| SQSTM1-213 | ENST00000510187 | 1714 | 378aa | ENSP00000424477 |
| SQSTM1-201 | ENST00000360718 | 2253 | 356aa | ENSP00000353944 |
| SQSTM1-208 | ENST00000485412 | 656 | No protein | - |
| SQSTM1-206 | ENST00000466342 | 808 | No protein | - |
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
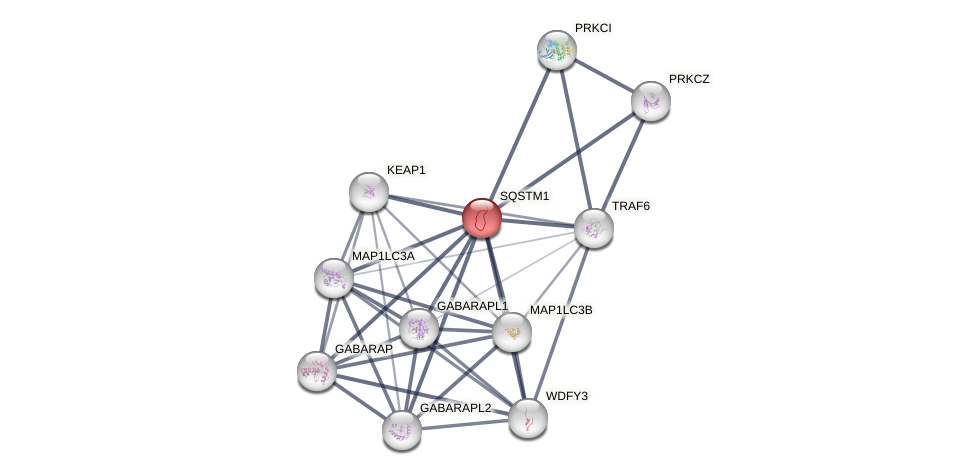
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000161011 | homo_sapiens | ENSP00000491834 | 100 | 100 | homo_sapiens | ENSP00000374455 | 100 | 100 |
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 44.3182 | 43.8636 | nomascus_leucogenys | ENSNLEP00000002152 | 98.9848 | 97.9695 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 99.7727 | 99.7727 | pan_paniscus | ENSPPAP00000022246 | 99.7727 | 99.7727 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 99.0909 | 98.1818 | pongo_abelii | ENSPPYP00000034065 | 99.0909 | 98.1818 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 99.7727 | 99.7727 | pan_troglodytes | ENSPTRP00000047790 | 99.7727 | 99.7727 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 99.7727 | 99.3182 | gorilla_gorilla | ENSGGOP00000020182 | 99.7727 | 99.3182 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 78.6364 | 67.5 | ornithorhynchus_anatinus | ENSOANP00000023386 | 75.3813 | 64.7059 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 68.8636 | 58.8636 | sarcophilus_harrisii | ENSSHAP00000028036 | 73.012 | 62.4096 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 56.3636 | 48.4091 | notamacropus_eugenii | ENSMEUP00000011407 | 72.093 | 61.9186 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 37.0455 | 34.0909 | choloepus_hoffmanni | ENSCHOP00000010066 | 48.368 | 44.5104 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 93.6364 | 89.5455 | dasypus_novemcinctus | ENSDNOP00000027378 | 93.2127 | 89.1403 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 77.0455 | 73.1818 | erinaceus_europaeus | ENSEEUP00000014532 | 90.8847 | 86.3271 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 59.7727 | 51.8182 | echinops_telfairi | ENSETEP00000015971 | 79.2169 | 68.6747 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 75.2273 | 69.7727 | echinops_telfairi | ENSETEP00000004412 | 89.7019 | 83.1978 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 97.5 | 96.1364 | callithrix_jacchus | ENSCJAP00000018685 | 97.7221 | 96.3554 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 98.8636 | 97.7273 | cercocebus_atys | ENSCATP00000021260 | 98.8636 | 97.7273 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 98.8636 | 97.7273 | macaca_fascicularis | ENSMFAP00000052246 | 93.3476 | 92.2747 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 98.6364 | 97.5 | macaca_mulatta | ENSMMUP00000033598 | 98.6364 | 97.5 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 99.0909 | 97.9545 | macaca_nemestrina | ENSMNEP00000004792 | 99.0909 | 97.9545 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 74.0909 | 73.1818 | papio_anubis | ENSPANP00000008739 | 99.3902 | 98.1707 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 83.8636 | 82.7273 | mandrillus_leucophaeus | ENSMLEP00000026690 | 98.6631 | 97.3262 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 94.7727 | 92.2727 | tursiops_truncatus | ENSTTRP00000004172 | 94.7727 | 92.2727 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 88.4091 | 84.0909 | vulpes_vulpes | ENSVVUP00000002031 | 87.6126 | 83.3333 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 95 | 91.1364 | mus_spretus | MGP_SPRETEiJ_P0027790 | 94.5701 | 90.724 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 95.9091 | 93.4091 | equus_asinus | ENSEASP00005018351 | 95.9091 | 93.4091 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 94.5455 | 93.4091 | capra_hircus | ENSCHIP00000026704 | 94.5455 | 93.4091 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 87.0455 | 83.8636 | loxodonta_africana | ENSLAFP00000027151 | 86.0674 | 82.9213 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 94.7727 | 92.5 | panthera_leo | ENSPLOP00000002644 | 79.2776 | 77.3764 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 84.7727 | 81.1364 | ailuropoda_melanoleuca | ENSAMEP00000034994 | 75.9674 | 72.7088 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 85 | 83.1818 | bos_indicus_hybrid | ENSBIXP00005000889 | 70.6994 | 69.1871 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 95.2273 | 92.7273 | oryctolagus_cuniculus | ENSOCUP00000013263 | 80.888 | 78.7645 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 95.6818 | 93.1818 | equus_caballus | ENSECAP00000018502 | 78.3985 | 76.3501 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 92.9545 | 90.2273 | canis_lupus_familiaris | ENSCAFP00845010418 | 88.3369 | 85.7451 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 94.7727 | 92.5 | panthera_pardus | ENSPPRP00000011179 | 93.7079 | 91.4607 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 95.9091 | 92.7273 | marmota_marmota_marmota | ENSMMMP00000011745 | 95.9091 | 92.7273 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 94.7727 | 92.5 | balaenoptera_musculus | ENSBMSP00010011380 | 94.7727 | 92.5 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 77.2727 | 72.2727 | cavia_porcellus | ENSCPOP00000023537 | 81.9277 | 76.6265 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 94.5455 | 92.2727 | felis_catus | ENSFCAP00000013637 | 93.9052 | 91.6479 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 84.7727 | 80.9091 | ursus_americanus | ENSUAMP00000035933 | 75.9674 | 72.5051 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 76.5909 | 65.9091 | monodelphis_domestica | ENSMODP00000046953 | 75.9009 | 65.3153 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 75.2273 | 74.0909 | bison_bison_bison | ENSBBBP00000004466 | 92.9775 | 91.573 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 95 | 92.5 | monodon_monoceros | ENSMMNP00015003734 | 95 | 92.5 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 95.4545 | 92.9545 | sus_scrofa | ENSSSCP00000056985 | 95.4545 | 92.9545 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 95.4545 | 92.7273 | microcebus_murinus | ENSMICP00000005976 | 95.4545 | 92.7273 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 87.7273 | 81.8182 | octodon_degus | ENSODEP00000010079 | 87.3303 | 81.448 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 78.4091 | 73.8636 | jaculus_jaculus | ENSJJAP00000015678 | 87.5635 | 82.4873 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 94.5455 | 90.6818 | jaculus_jaculus | ENSJJAP00000007715 | 94.5455 | 90.6818 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 75.9091 | 70.6818 | jaculus_jaculus | ENSJJAP00000011199 | 84.9873 | 79.1349 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 93.4091 | 92.2727 | ovis_aries_rambouillet | ENSOARP00020018175 | 93.8356 | 92.6941 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 52.0455 | 46.3636 | ursus_maritimus | ENSUMAP00000033633 | 58.5678 | 52.1739 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 62.0455 | 57.7273 | ochotona_princeps | ENSOPRP00000014198 | 62.9032 | 58.5253 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 95 | 92.5 | delphinapterus_leucas | ENSDLEP00000023403 | 95 | 92.5 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 94.5455 | 92.2727 | physeter_catodon | ENSPCTP00005001705 | 94.5455 | 92.2727 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 94.5455 | 91.1364 | mus_caroli | MGP_CAROLIEiJ_P0026424 | 94.1176 | 90.724 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 94.7727 | 91.1364 | mus_musculus | ENSMUSP00000099835 | 94.3439 | 90.724 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 85 | 80.2273 | dipodomys_ordii | ENSDORP00000013972 | 86.3741 | 81.5242 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 77.9545 | 75.2273 | mustela_putorius_furo | ENSMPUP00000000647 | 92.4528 | 89.2183 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 98.6364 | 96.5909 | aotus_nancymaae | ENSANAP00000037667 | 98.6364 | 96.5909 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 97.5 | 95.6818 | saimiri_boliviensis_boliviensis | ENSSBOP00000015157 | 97.7221 | 95.8998 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 41.5909 | 40.4545 | vicugna_pacos | ENSVPAP00000007276 | 64.8936 | 63.1206 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 98.4091 | 96.5909 | chlorocebus_sabaeus | ENSCSAP00000004594 | 98.4091 | 96.5909 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 95.9091 | 92.2727 | mesocricetus_auratus | ENSMAUP00000014683 | 95.4751 | 91.8552 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 94.7727 | 92.2727 | phocoena_sinus | ENSPSNP00000012505 | 94.7727 | 92.2727 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 95.2273 | 93.4091 | camelus_dromedarius | ENSCDRP00005007725 | 95.2273 | 93.4091 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 76.1364 | 73.1818 | carlito_syrichta | ENSTSYP00000013720 | 90.5405 | 87.027 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 76.1364 | 73.8636 | panthera_tigris_altaica | ENSPTIP00000025692 | 94.3662 | 91.5493 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 94.5455 | 91.3636 | rattus_norvegicus | ENSRNOP00000056021 | 94.7608 | 91.5718 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 95.4545 | 93.4091 | prolemur_simus | ENSPSMP00000015733 | 95.672 | 93.6219 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 95 | 91.8182 | peromyscus_maniculatus_bairdii | ENSPEMP00000021719 | 94.5701 | 91.4027 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 75 | 73.8636 | bos_mutus | ENSBMUP00000019716 | 92.6966 | 91.2921 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 95 | 91.1364 | mus_spicilegus | ENSMSIP00000010952 | 93.722 | 89.9103 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 95.9091 | 92.0455 | cricetulus_griseus_chok1gshd | ENSCGRP00001007259 | 95.4751 | 91.629 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 83.8636 | 80.6818 | pteropus_vampyrus | ENSPVAP00000014615 | 83.6735 | 80.4989 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 78.6364 | 75 | myotis_lucifugus | ENSMLUP00000001147 | 93.2615 | 88.9488 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 77.5 | 67.0455 | vombatus_ursinus | ENSVURP00010025679 | 76.6292 | 66.2921 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 94.7727 | 92.2727 | rhinolophus_ferrumequinum | ENSRFEP00010032196 | 94.3439 | 91.8552 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 74.3182 | 71.5909 | neovison_vison | ENSNVIP00000012560 | 92.6346 | 89.2351 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 81.1364 | 77.7273 | bos_taurus | ENSBTAP00000059264 | 68.26 | 65.392 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 82.5 | 78.4091 | heterocephalus_glaber_female | ENSHGLP00000056261 | 71.3163 | 67.78 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 97.7273 | 96.3636 | rhinopithecus_bieti | ENSRBIP00000033343 | 95.3437 | 94.0133 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 92.9545 | 90.2273 | canis_lupus_dingo | ENSCAFP00020003110 | 92.3251 | 89.6162 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 96.5909 | 93.6364 | sciurus_vulgaris | ENSSVLP00005005785 | 96.5909 | 93.6364 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 77.2727 | 66.3636 | phascolarctos_cinereus | ENSPCIP00000004759 | 61.3718 | 52.7076 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 94.3182 | 92.0455 | procavia_capensis | ENSPCAP00000002271 | 94.7489 | 92.4658 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 94.5455 | 91.1364 | nannospalax_galili | ENSNGAP00000000129 | 94.5455 | 91.1364 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 84.7727 | 82.9545 | bos_grunniens | ENSBGRP00000007488 | 70.5104 | 68.9981 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 83.6364 | 78.8636 | chinchilla_lanigera | ENSCLAP00000021716 | 74.3434 | 70.101 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 94.5455 | 92.2727 | cervus_hanglu_yarkandensis | ENSCHYP00000036985 | 94.5455 | 92.2727 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 95.4545 | 92.5 | catagonus_wagneri | ENSCWAP00000000274 | 95.4545 | 92.5 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 96.3636 | 93.4091 | ictidomys_tridecemlineatus | ENSSTOP00000032458 | 96.3636 | 93.4091 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 95.4545 | 93.1818 | otolemur_garnettii | ENSOGAP00000001924 | 94.1704 | 91.9283 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 97.5 | 96.3636 | cebus_imitator | ENSCCAP00000032336 | 97.7221 | 96.5831 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 96.1364 | 92.9545 | urocitellus_parryii | ENSUPAP00010016282 | 96.1364 | 92.9545 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 94.5455 | 92.5 | moschus_moschiferus | ENSMMSP00000019919 | 94.5455 | 92.5 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 97.7273 | 96.3636 | rhinopithecus_roxellana | ENSRROP00000024677 | 95.3437 | 94.0133 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 94.7727 | 90.9091 | mus_pahari | MGP_PahariEiJ_P0033200 | 94.3439 | 90.4977 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 93.1818 | 89.5455 | propithecus_coquereli | ENSPCOP00000015269 | 93.1818 | 89.5455 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 18.4091 | 11.1364 | caenorhabditis_elegans | Y40C5A.1.1 | 33.4711 | 20.2479 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 31.3636 | 19.5455 | caenorhabditis_elegans | C06G3.6.2 | 27.7108 | 17.2691 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 40 | 20.6818 | caenorhabditis_elegans | T12G3.1a.1 | 25.3968 | 13.1313 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 13.6364 | 8.63636 | caenorhabditis_elegans | Y32B12A.1.1 | 33.8983 | 21.4689 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 15.4545 | 10.2273 | caenorhabditis_elegans | T26H2.5.1 | 29.6943 | 19.6507 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 42.0455 | 24.3182 | drosophila_melanogaster | FBpp0099683 | 30.8848 | 17.8631 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 20.2273 | 12.0455 | ciona_intestinalis | ENSCINP00000033899 | 47.8495 | 28.4946 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 34.0909 | 24.3182 | ciona_intestinalis | ENSCINP00000023991 | 45.5927 | 32.5228 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 22.9545 | 14.0909 | ciona_intestinalis | ENSCINP00000002565 | 45.0893 | 27.6786 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 46.8182 | 34.7727 | petromyzon_marinus | ENSPMAP00000003656 | 45.3745 | 33.7004 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 56.5909 | 44.3182 | callorhinchus_milii | ENSCMIP00000013654 | 57.1101 | 44.7248 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 63.1818 | 47.2727 | latimeria_chalumnae | ENSLACP00000013831 | 63.0385 | 47.1655 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 60.2273 | 46.1364 | lepisosteus_oculatus | ENSLOCP00000014082 | 60.2273 | 46.1364 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 60.4545 | 47.5 | clupea_harengus | ENSCHAP00000045127 | 57.8261 | 45.4348 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 60.9091 | 47.0455 | danio_rerio | ENSDARP00000117113 | 59.292 | 45.7965 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 58.6364 | 44.3182 | carassius_auratus | ENSCARP00000101827 | 50.2924 | 38.0117 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 61.3636 | 47.5 | carassius_auratus | ENSCARP00000004452 | 59.2105 | 45.8333 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 59.0909 | 45.9091 | astyanax_mexicanus | ENSAMXP00000011836 | 60.8899 | 47.3068 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 60.6818 | 47.9545 | ictalurus_punctatus | ENSIPUP00000019226 | 58.0435 | 45.8696 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 59.3182 | 45.9091 | esox_lucius | ENSELUP00000022664 | 55.2966 | 42.7966 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 59.0909 | 47.7273 | electrophorus_electricus | ENSEEEP00000050660 | 57.0175 | 46.0526 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 59.0909 | 45.9091 | oncorhynchus_kisutch | ENSOKIP00005012001 | 55.4371 | 43.0704 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 58.4091 | 45.4545 | oncorhynchus_kisutch | ENSOKIP00005087766 | 55.3879 | 43.1034 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 58.8636 | 45.6818 | oncorhynchus_mykiss | ENSOMYP00000099051 | 55.2239 | 42.8571 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 49.0909 | 37.7273 | oncorhynchus_mykiss | ENSOMYP00000111587 | 44.5361 | 34.2268 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 58.1818 | 45 | salmo_salar | ENSSSAP00000104849 | 54.1226 | 41.8605 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 58.6364 | 45.4545 | salmo_salar | ENSSSAP00000097368 | 56.087 | 43.4783 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 58.1818 | 44.7727 | salmo_trutta | ENSSTUP00000089044 | 55.2916 | 42.5486 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 58.8636 | 45.4545 | salmo_trutta | ENSSTUP00000062521 | 55.2239 | 42.6439 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 58.4091 | 46.8182 | gadus_morhua | ENSGMOP00000025600 | 57.6233 | 46.1883 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 59.5455 | 48.1818 | fundulus_heteroclitus | ENSFHEP00000009045 | 58.2222 | 47.1111 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 58.8636 | 47.2727 | poecilia_reticulata | ENSPREP00000016733 | 51.8 | 41.6 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 57.0455 | 45.9091 | oryzias_latipes | ENSORLP00000025483 | 57.5688 | 46.3303 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 57.7273 | 44.5455 | cyclopterus_lumpus | ENSCLMP00005007680 | 60.7655 | 46.89 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 58.8636 | 46.5909 | oreochromis_niloticus | ENSONIP00000000126 | 59.4037 | 47.0183 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 58.8636 | 46.5909 | haplochromis_burtoni | ENSHBUP00000015745 | 60.7981 | 48.1221 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 59.0909 | 46.8182 | astatotilapia_calliptera | ENSACLP00000027648 | 60.7477 | 48.1308 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 42.7273 | 31.8182 | lates_calcarifer | ENSLCAP00010000923 | 55.132 | 41.0557 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 69.7727 | 55.9091 | xenopus_tropicalis | ENSXETP00000034720 | 67.3246 | 53.9474 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 70.9091 | 59.3182 | chrysemys_picta_bellii | ENSCPBP00000012733 | 69.0266 | 57.7434 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 71.5909 | 60 | sphenodon_punctatus | ENSSPUP00000016116 | 66.5962 | 55.814 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 49.3182 | 37.7273 | crocodylus_porosus | ENSCPRP00005016854 | 60.6145 | 46.3687 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 67.0455 | 55.9091 | laticauda_laticaudata | ENSLLTP00000018399 | 58.3004 | 48.6166 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 67.0455 | 55.9091 | notechis_scutatus | ENSNSUP00000000087 | 65.5556 | 54.6667 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 67.0455 | 55.6818 | pseudonaja_textilis | ENSPTXP00000019508 | 65.5556 | 54.4444 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 72.2727 | 60.6818 | gallus_gallus | ENSGALP00010023771 | 70.1987 | 58.9404 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 60.6818 | 50.6818 | meleagris_gallopavo | ENSMGAP00000007267 | 70.6349 | 58.9947 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 71.1364 | 59.5455 | serinus_canaria | ENSSCAP00000015769 | 69.0949 | 57.8366 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 71.8182 | 60.4545 | parus_major | ENSPMJP00000010384 | 52.7546 | 44.4073 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 58.6364 | 46.5909 | anolis_carolinensis | ENSACAP00000021254 | 63.2353 | 50.2451 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 59.0909 | 46.5909 | neolamprologus_brichardi | ENSNBRP00000001286 | 60.7477 | 47.8972 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 66.1364 | 55 | naja_naja | ENSNNAP00000022901 | 64.0969 | 53.304 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 68.4091 | 57.2727 | podarcis_muralis | ENSPMRP00000009451 | 67.1875 | 56.25 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 60 | 50.6818 | geospiza_fortis | ENSGFOP00000000011 | 70.2128 | 59.3085 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 71.5909 | 60 | taeniopygia_guttata | ENSTGUP00000014177 | 69.5364 | 58.2781 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 61.1364 | 48.1818 | erpetoichthys_calabaricus | ENSECRP00000015000 | 63.5934 | 50.1182 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 58.6364 | 46.3636 | kryptolebias_marmoratus | ENSKMAP00000024409 | 60 | 47.4419 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 68.4091 | 55.9091 | salvator_merianae | ENSSMRP00000022155 | 66.0088 | 53.9474 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 57.2727 | 45.4545 | oryzias_javanicus | ENSOJAP00000003560 | 61.3139 | 48.6618 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 60.4545 | 47.5 | amphilophus_citrinellus | ENSACIP00000012626 | 60.8696 | 47.8261 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 59.0909 | 46.1364 | dicentrarchus_labrax | ENSDLAP00005079230 | 60.8899 | 47.541 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 39.7727 | 31.3636 | amphiprion_percula | ENSAPEP00000007432 | 59.322 | 46.7797 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 16.5909 | 12.0455 | amphiprion_percula | ENSAPEP00000007389 | 64.6018 | 46.9027 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 67.7273 | 56.1364 | pelodiscus_sinensis | ENSPSIP00000010714 | 68.1922 | 56.5217 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 71.3636 | 59.3182 | gopherus_evgoodei | ENSGEVP00005006194 | 68.709 | 57.1116 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 71.5909 | 59.5455 | chelonoidis_abingdonii | ENSCABP00000017874 | 68.9278 | 57.3304 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 59.5455 | 46.5909 | seriola_dumerili | ENSSDUP00000022357 | 63.4383 | 49.6368 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 34.0909 | 26.8182 | cottoperca_gobio | ENSCGOP00000048363 | 66.6667 | 52.4444 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 65.2273 | 55.2273 | struthio_camelus_australis | ENSSCUP00000003907 | 72.1105 | 61.0553 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 72.0455 | 62.0455 | anser_brachyrhynchus | ENSABRP00000021514 | 74.0654 | 63.785 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 58.6364 | 45.2273 | gasterosteus_aculeatus | ENSGACP00000021468 | 61.2827 | 47.2684 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 54.3182 | 44.0909 | cynoglossus_semilaevis | ENSCSEP00000008395 | 55.9719 | 45.4333 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 58.8636 | 47.5 | poecilia_formosa | ENSPFOP00000017211 | 55.9395 | 45.1404 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 60 | 48.4091 | myripristis_murdjan | ENSMMDP00005031993 | 54.2094 | 43.7372 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 58.6364 | 45.4545 | sander_lucioperca | ENSSLUP00000050444 | 60.5634 | 46.9484 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 72.5 | 60.4545 | strigops_habroptila | ENSSHBP00005009678 | 70.5752 | 58.8496 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 59.5455 | 46.8182 | amphiprion_ocellaris | ENSAOCP00000030674 | 61.9385 | 48.6998 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 70.9091 | 58.8636 | terrapene_carolina_triunguis | ENSTMTP00000030343 | 68.8742 | 57.1744 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 58.1818 | 44.7727 | hucho_hucho | ENSHHUP00000026670 | 54.9356 | 42.2747 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 59.3182 | 46.1364 | hucho_hucho | ENSHHUP00000001229 | 57.6159 | 44.8124 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 56.8182 | 46.5909 | betta_splendens | ENSBSLP00000016856 | 57.0776 | 46.8037 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 61.3636 | 48.6364 | pygocentrus_nattereri | ENSPNAP00000031921 | 57.9399 | 45.9227 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 59.3182 | 48.1818 | anabas_testudineus | ENSATEP00000028099 | 62.8916 | 51.0843 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 35.6818 | 22.9545 | ciona_savignyi | ENSCSAVP00000008508 | 48.1595 | 30.9816 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 59.5455 | 46.3636 | seriola_lalandi_dorsalis | ENSSLDP00000021742 | 60.7889 | 47.3318 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 17.2727 | 11.3636 | labrus_bergylta | ENSLBEP00000032673 | 70.3704 | 46.2963 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 38.4091 | 29.7727 | labrus_bergylta | ENSLBEP00000032661 | 62.1324 | 48.1618 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 59.0909 | 46.8182 | pundamilia_nyererei | ENSPNYP00000000010 | 60.7477 | 48.1308 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 58.4091 | 46.1364 | mastacembelus_armatus | ENSMAMP00000050286 | 60.6132 | 47.8774 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 60.6818 | 48.1818 | stegastes_partitus | ENSSPAP00000025556 | 64.3373 | 51.0843 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 54.3182 | 41.5909 | oncorhynchus_tshawytscha | ENSOTSP00005094857 | 50.9595 | 39.0192 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 58.1818 | 45.4545 | oncorhynchus_tshawytscha | ENSOTSP00005098606 | 56.3877 | 44.0529 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 57.9545 | 45.2273 | acanthochromis_polyacanthus | ENSAPOP00000016273 | 62.6536 | 48.8943 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 73.4091 | 61.5909 | aquila_chrysaetos_chrysaetos | ENSACCP00020021841 | 71.3024 | 59.8234 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 57.2727 | 46.1364 | nothobranchius_furzeri | ENSNFUP00015005016 | 59.5745 | 47.9905 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 59.3182 | 47.7273 | cyprinodon_variegatus | ENSCVAP00000022201 | 60 | 48.2759 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 61.5909 | 47.5 | denticeps_clupeoides | ENSDCDP00000019108 | 56.341 | 43.4511 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 58.1818 | 46.1364 | larimichthys_crocea | ENSLCRP00005009302 | 51.0978 | 40.519 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 57.7273 | 46.3636 | takifugu_rubripes | ENSTRUP00000081395 | 55.4585 | 44.5415 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 67.0455 | 56.5909 | ficedula_albicollis | ENSFALP00000016530 | 62.6327 | 52.8662 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 53.8636 | 42.9545 | hippocampus_comes | ENSHCOP00000010793 | 58.231 | 46.4373 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 58.6364 | 43.8636 | cyprinus_carpio_carpio | ENSCCRP00000018012 | 53.8622 | 40.2923 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 60.9091 | 46.8182 | cyprinus_carpio_carpio | ENSCCRP00000121651 | 58.0087 | 44.5887 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 68.8636 | 59.3182 | coturnix_japonica | ENSCJPP00005005690 | 68.3973 | 58.9165 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 44.3182 | 31.8182 | sinocyclocheilus_grahami | ENSSGRP00000004976 | 61.1285 | 43.8871 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 14.0909 | 10.4545 | sinocyclocheilus_grahami | ENSSGRP00000004972 | 68.8889 | 51.1111 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 14.0909 | 10.4545 | sinocyclocheilus_grahami | ENSSGRP00000086244 | 68.8889 | 51.1111 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 60.9091 | 46.5909 | sinocyclocheilus_grahami | ENSSGRP00000015322 | 56.3025 | 43.0672 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 59.0909 | 46.8182 | maylandia_zebra | ENSMZEP00005029599 | 60.7477 | 48.1308 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 55.2273 | 45.6818 | tetraodon_nigroviridis | ENSTNIP00000005606 | 57.7197 | 47.7435 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 58.4091 | 45 | paramormyrops_kingsleyae | ENSPKIP00000036790 | 61.0451 | 47.0309 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 59.5455 | 47.0455 | scleropages_formosus | ENSSFOP00015004346 | 59.4104 | 46.9388 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 68.6364 | 55 | leptobrachium_leishanense | ENSLLEP00000031064 | 52.4306 | 42.0139 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 62.2727 | 49.5455 | leptobrachium_leishanense | ENSLLEP00000029675 | 65.5502 | 52.1531 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 56.5909 | 46.3636 | scophthalmus_maximus | ENSSMAP00000046075 | 61.33 | 50.2463 |
| ENSG00000161011 | homo_sapiens | ENSP00000374455 | 58.1818 | 46.5909 | oryzias_melastigma | ENSOMEP00000006663 | 57.9186 | 46.3801 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0000122 | involved_in negative regulation of transcription by RNA polymerase II | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0000407 | located_in phagophore assembly site | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0000422 | involved_in autophagy of mitochondrion | 1 | NAS | Homo_sapiens(9606) | Process |
| GO:0000423 | involved_in mitophagy | 2 | IBA,IGI | Homo_sapiens(9606) | Process |
| GO:0000425 | involved_in pexophagy | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0000932 | located_in P-body | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0001659 | involved_in temperature homeostasis | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0001934 | involved_in positive regulation of protein phosphorylation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0002376 | involved_in immune system process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0002931 | involved_in response to ischemia | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0004674 | enables protein serine/threonine kinase activity | 1 | NAS | Homo_sapiens(9606) | Function |
| GO:0005080 | enables protein kinase C binding | 2 | IBA,IPI | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005654 | located_in nucleoplasm | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0005737 | located_in cytoplasm | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005739 | located_in mitochondrion | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0005770 | located_in late endosome | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0005776 | is_active_in autophagosome | 2 | IDA | Homo_sapiens(9606) | Component |
| GO:0005783 | located_in endoplasmic reticulum | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0005829 | located_in cytosol | 2 | IDA,TAS | Homo_sapiens(9606) | Component |
| GO:0006366 | involved_in transcription by RNA polymerase II | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0006468 | involved_in protein phosphorylation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0006511 | involved_in ubiquitin-dependent protein catabolic process | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0006606 | involved_in protein import into nucleus | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0006914 | involved_in autophagy | 3 | IDA,IMP,TAS | Homo_sapiens(9606) | Process |
| GO:0006915 | involved_in apoptotic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0007032 | involved_in endosome organization | 2 | IBA,IDA | Homo_sapiens(9606) | Process |
| GO:0008104 | involved_in protein localization | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0008270 | enables zinc ion binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0010508 | involved_in positive regulation of autophagy | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0010821 | involved_in regulation of mitochondrion organization | 1 | NAS | Homo_sapiens(9606) | Process |
| GO:0016197 | involved_in endosomal transport | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0016234 | located_in inclusion body | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0016235 | is_active_in aggresome | 1 | IBA | Homo_sapiens(9606) | Component |
| GO:0016236 | involved_in macroautophagy | 2 | IMP,ISS | Homo_sapiens(9606) | Process |
| GO:0016605 | located_in PML body | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0019899 | enables enzyme binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0019901 | enables protein kinase binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0030017 | located_in sarcomere | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0030154 | involved_in cell differentiation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0030163 | involved_in protein catabolic process | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0030674 | enables protein-macromolecule adaptor activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0030971 | enables receptor tyrosine kinase binding | 1 | TAS | Homo_sapiens(9606) | Function |
| GO:0031397 | involved_in negative regulation of protein ubiquitination | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0031625 | enables ubiquitin protein ligase binding | 2 | IDA,IPI | Homo_sapiens(9606) | Function |
| GO:0034614 | involved_in cellular response to reactive oxygen species | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0035255 | enables ionotropic glutamate receptor binding | 1 | ISS | Homo_sapiens(9606) | Function |
| GO:0035556 | involved_in intracellular signal transduction | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0035591 | enables signaling adaptor activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0035973 | involved_in aggrephagy | 2 | IBA,IPI | Homo_sapiens(9606) | Process |
| GO:0038023 | enables signaling receptor activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0042169 | enables SH2 domain binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0042802 | enables identical protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0043065 | involved_in positive regulation of apoptotic process | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0043122 | involved_in regulation of I-kappaB kinase/NF-kappaB signaling | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0043130 | enables ubiquitin binding | 2 | IDA,TAS | Homo_sapiens(9606) | Function |
| GO:0043231 | located_in intracellular membrane-bounded organelle | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0044753 | is_active_in amphisome | 2 | IBA,IDA | Homo_sapiens(9606) | Component |
| GO:0044754 | located_in autolysosome | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0044877 | enables protein-containing complex binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0045944 | involved_in positive regulation of transcription by RNA polymerase II | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0046578 | involved_in regulation of Ras protein signal transduction | 1 | NAS | Homo_sapiens(9606) | Process |
| GO:0061635 | involved_in regulation of protein complex stability | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0061912 | involved_in selective autophagy | 2 | IDA,IMP | Homo_sapiens(9606) | Process |
| GO:0070062 | located_in extracellular exosome | 1 | HDA | Homo_sapiens(9606) | Component |
| GO:0070342 | involved_in brown fat cell proliferation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0070530 | enables K63-linked polyubiquitin modification-dependent protein binding | 2 | IBA,IDA | Homo_sapiens(9606) | Function |
| GO:0071211 | involved_in protein targeting to vacuole involved in autophagy | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0097009 | involved_in energy homeostasis | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0097225 | located_in sperm midpiece | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0097413 | located_in Lewy body | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0098780 | involved_in response to mitochondrial depolarisation | 1 | IGI | Homo_sapiens(9606) | Process |
| GO:0140036 | enables ubiquitin-dependent protein binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:1900273 | involved_in positive regulation of long-term synaptic potentiation | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:1903078 | involved_in positive regulation of protein localization to plasma membrane | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:1905719 | involved_in protein localization to perinuclear region of cytoplasm | 1 | IDA | Homo_sapiens(9606) | Process |