Welcome to
RBP World!
Functions and Diseases
| RBP Type | Canonical_RBPs |
| Diseases | Cancer Disease |
| Drug | N.A. |
| Main interacting RNAs | diverse |
| Moonlighting functions | N.A. |
| Localizations | Stress granucle |
| BulkPerturb-seq | DataSet_01_241 DataSet_01_375 |
Description
| Ensembl ID | ENSG00000138035 | Gene ID | 87178 | Accession | 23166 |
| Symbol | PNPT1 | Alias | OLD35;SCA25;DFNB70;PNPASE;old-35;COXPD13 | Full Name | polyribonucleotide nucleotidyltransferase 1 |
| Status | Confidence | Length | 59803 bases | Strand | Minus strand |
| Position | 2 : 55634061 - 55693863 | RNA binding domain | RNase_PH , RNase_PH_C , PNPase , KH_1 | ||
| Summary | The protein encoded by this gene belongs to the evolutionary conserved polynucleotide phosphorylase family comprised of phosphate dependent 3'-to-5' exoribonucleases implicated in RNA processing and degradation. This enzyme is predominantly localized in the mitochondrial intermembrane space and is involved in import of RNA to mitochondria. Mutations in this gene have been associated with combined oxidative phosphorylation deficiency-13 and autosomal recessive nonsyndromic deafness-70. Related pseudogenes are found on chromosomes 3 and 7. [provided by RefSeq, Dec 2012] | ||||
RNA binding domains (RBDs)
| Protein | Domain | Pfam ID | E-value | Domain number |
|---|---|---|---|---|
| ENSP00000393953 | RNase_PH_C | PF03725 | 1e-16 | 1 |
| ENSP00000393953 | KH_1 | PF00013 | 1.9e-07 | 2 |
| ENSP00000393953 | PNPase | PF03726 | 2.7e-07 | 3 |
| ENSP00000393953 | RNase_PH | PF01138 | 3.7e-39 | 4 |
| ENSP00000393953 | RNase_PH | PF01138 | 3.7e-39 | 5 |
| ENSP00000400646 | RNase_PH_C | PF03725 | 1e-16 | 1 |
| ENSP00000400646 | KH_1 | PF00013 | 1.9e-07 | 2 |
| ENSP00000400646 | PNPase | PF03726 | 2.7e-07 | 3 |
| ENSP00000400646 | RNase_PH | PF01138 | 3.7e-39 | 4 |
| ENSP00000400646 | RNase_PH | PF01138 | 3.7e-39 | 5 |
| ENSP00000260604 | RNase_PH | PF01138 | 9.9e-13 | 1 |
| ENSP00000411057 | RNase_PH | PF01138 | 6.4e-07 | 1 |
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|---|---|---|---|
| 32574647 | Regulation of cellular senescence by eukaryotic members of the FAH superfamily - A role in calcium homeostasis? | H K Alexander Weiss | 2020-09-01 | Mechanisms of ageing and development |
| 32809827 | A Novel Redox Modulator Induces a GPX4-Mediated Cell Death That Is Dependent on Iron and Reactive Oxygen Species. | Shuai Hu | 2020-09-10 | Journal of medicinal chemistry |
| 32221480 | Analysis of mitochondrial m1A/G RNA modification reveals links to nuclear genetic variants and associated disease processes. | Tasnim Aminah Ali | 2020-03-27 | Communications biology |
| 33199448 | Heterogeneity of PNPT1 neuroimaging: mitochondriopathy, interferonopathy or both? | Alessandra Pennisi | 2022-02-01 | Journal of medical genetics |
| 36266447 | 8-Oxoguanine: from oxidative damage to epigenetic and epitranscriptional modification. | Young Ja Hahm | 2022-10-01 | Experimental & molecular medicine |
| 28645153 | Defective mitochondrial RNA processing due to PNPT1 variants causes Leigh syndrome. | Sanna Matilainen | 2017-09-01 | Human molecular genetics |
| 34074335 | Circ-PNPT1 contributes to gestational diabetes mellitus (GDM) by regulating the function of trophoblast cells through miR-889-3p/PAK1 axis. | Li Zhang | 2021-06-01 | Diabetology & metabolic syndrome |
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| PNPT1-205 | ENST00000447944 | 4549 | 783aa | ENSP00000400646 |
| PNPT1-202 | ENST00000415374 | 3308 | 783aa | ENSP00000393953 |
| PNPT1-201 | ENST00000260604 | 3596 | 179aa | ENSP00000260604 |
| PNPT1-206 | ENST00000481066 | 1281 | No protein | - |
| PNPT1-203 | ENST00000415489 | 555 | 139aa | ENSP00000411057 |
| PNPT1-204 | ENST00000429805 | 583 | 71aa | ENSP00000411994 |
| PNPT1-207 | ENST00000625249 | 216 | 71aa | ENSP00000486227 |
Phenotypes
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| 15318 | Heart Failure | 2.9347593E-004 | - |
| 17450 | Blood Pressure | 5.821e-006 | 19060910 |
| 17451 | Blood Pressure | 2.677e-005 | 19060910 |
| 17452 | Blood Pressure | 9.378e-006 | 19060910 |
| 17467 | Blood Pressure | 1.4e-005 | 19060910 |
| 25161 | Body Weight | 2.6980000E-005 | - |
| 25177 | Body Weight | 2.1730000E-005 | - |
| 25178 | Body Weight | 7.5780000E-005 | - |
| 25199 | Body Weight | 5.4190000E-005 | - |
| 28431 | Nutritional and Metabolic Diseases | 2.6640000E-005 | - |
| 29438 | Cholesterol, LDL | 4.9860000E-005 | - |
| 41205 | Platelet Function Tests | 5.8360505E-006 | - |
| 46913 | Platelet Function Tests | 7.2638000E-006 | - |
| 48255 | Child Development Disorders, Pervasive | 4.7078500E-005 | - |
| 58650 | Respiratory Function Tests | 2.1665000E-008 | 21946350 |
| 58651 | Respiratory Function Tests | 2.2117000E-008 | 21946350 |
| 58652 | Respiratory Function Tests | 1.7882000E-008 | 21946350 |
| 58654 | Respiratory Function Tests | 2.1456000E-008 | 21946350 |
| 58655 | Respiratory Function Tests | 2.0981000E-008 | 21946350 |
| 74877 | Body Height | 4E-8 | 20881960 |
| 81379 | Amyotrophic Lateral Sclerosis | 1E-6 | 19451621 |
| 87171 | Immunoglobulins | 5E-6 | 23382691 |
| 98017 | Body Height | 1E-25 | 25429064 |
| 101623 | 3-hydroxy-1-methylpropylmercapturic acid | 4E-7 | 26053186 |
| 101624 | Smoking | 4E-7 | 26053186 |
| 103850 | Hernia, Inguinal | 1E-17 | 26686553 |
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
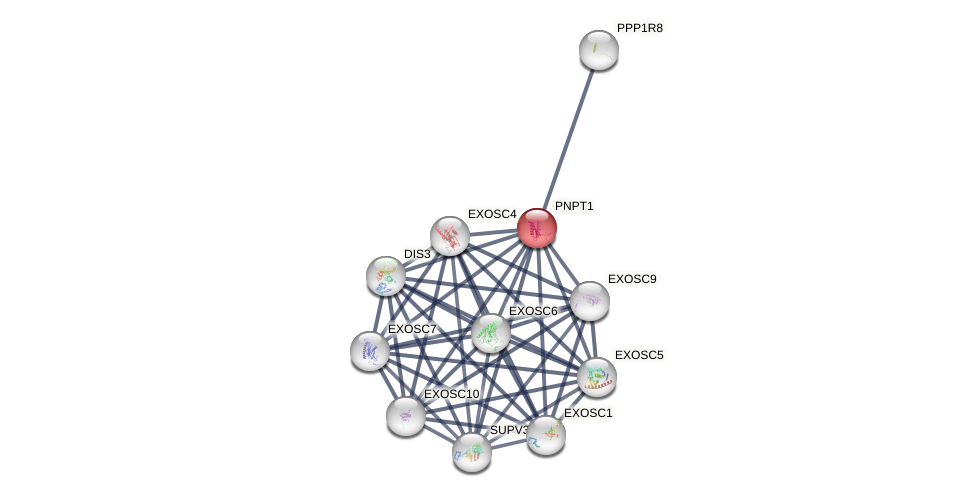
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000138035 | ||||||||
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 96.9349 | 95.6577 | nomascus_leucogenys | ENSNLEP00000011430 | 98.3161 | 97.0207 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 89.9106 | 89.7829 | pan_paniscus | ENSPPAP00000006254 | 99.7167 | 99.5751 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 89.5275 | 88.7612 | pongo_abelii | ENSPPYP00000013833 | 99.7155 | 98.862 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 98.9783 | 98.4674 | pan_troglodytes | ENSPTRP00000084156 | 98.852 | 98.3418 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 99.8723 | 99.6169 | gorilla_gorilla | ENSGGOP00000006726 | 99.8723 | 99.6169 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 91.6986 | 83.2695 | ornithorhynchus_anatinus | ENSOANP00000034632 | 87.6679 | 79.6093 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 93.742 | 84.9298 | sarcophilus_harrisii | ENSSHAP00000036157 | 93.5032 | 84.7134 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 79.8212 | 76.6283 | choloepus_hoffmanni | ENSCHOP00000006537 | 86.2069 | 82.7586 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 96.8072 | 92.0817 | dasypus_novemcinctus | ENSDNOP00000024986 | 96.6837 | 91.9643 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 87.6117 | 82.5032 | erinaceus_europaeus | ENSEEUP00000014260 | 87.1665 | 82.0839 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 73.8186 | 68.327 | echinops_telfairi | ENSETEP00000001279 | 73.7245 | 68.2398 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 98.212 | 96.0409 | callithrix_jacchus | ENSCJAP00000018027 | 98.0867 | 95.9184 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 97.5734 | 96.2963 | cercocebus_atys | ENSCATP00000015303 | 98.0745 | 96.7908 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 98.5951 | 97.5734 | macaca_fascicularis | ENSMFAP00000004449 | 98.5951 | 97.5734 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 98.7229 | 97.5734 | macaca_mulatta | ENSMMUP00000009189 | 98.7229 | 97.5734 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 93.4866 | 92.2094 | macaca_nemestrina | ENSMNEP00000030323 | 97.0822 | 95.756 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 92.4649 | 91.3155 | papio_anubis | ENSPANP00000023612 | 99.3141 | 98.0796 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 90.5492 | 89.272 | papio_anubis | ENSPANP00000059117 | 98.4722 | 97.0833 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 98.7229 | 97.7011 | papio_anubis | ENSPANP00000008826 | 98.7229 | 97.7011 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 95.4023 | 94.2529 | mandrillus_leucophaeus | ENSMLEP00000029076 | 98.5488 | 97.3615 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 90.6769 | 86.9732 | tursiops_truncatus | ENSTTRP00000000027 | 97.3937 | 93.4156 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 97.7011 | 93.4866 | vulpes_vulpes | ENSVVUP00000013674 | 97.5765 | 93.3673 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 96.424 | 91.4432 | mus_spretus | MGP_SPRETEiJ_P0027296 | 96.301 | 91.3265 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 97.8289 | 94.3806 | equus_asinus | ENSEASP00005047757 | 95.6305 | 92.2597 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 92.848 | 88.1226 | capra_hircus | ENSCHIP00000030286 | 95.5322 | 90.6702 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 91.3155 | 85.4406 | loxodonta_africana | ENSLAFP00000016925 | 91.0828 | 85.2229 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 51.2133 | 49.8084 | panthera_leo | ENSPLOP00000002176 | 98.044 | 95.3545 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 97.4457 | 93.9974 | ailuropoda_melanoleuca | ENSAMEP00000004040 | 97.3214 | 93.8775 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 96.424 | 91.6986 | bos_indicus_hybrid | ENSBIXP00005013589 | 96.424 | 91.6986 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 97.1903 | 93.8697 | oryctolagus_cuniculus | ENSOCUP00000005963 | 97.0663 | 93.75 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 97.8289 | 94.3806 | equus_caballus | ENSECAP00000018223 | 93.5287 | 90.232 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 97.5734 | 93.742 | canis_lupus_familiaris | ENSCAFP00845016683 | 97.449 | 93.6225 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 97.7011 | 94.636 | panthera_pardus | ENSPPRP00000005674 | 97.5765 | 94.5153 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 97.4457 | 93.1034 | marmota_marmota_marmota | ENSMMMP00000008191 | 97.3214 | 92.9847 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 95.6577 | 91.8263 | balaenoptera_musculus | ENSBMSP00010026141 | 96.5206 | 92.6546 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 97.1903 | 91.6986 | cavia_porcellus | ENSCPOP00000009894 | 97.1903 | 91.6986 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 97.5734 | 94.3806 | felis_catus | ENSFCAP00000012275 | 97.449 | 94.2602 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 84.802 | 81.3538 | ursus_americanus | ENSUAMP00000023753 | 95.6772 | 91.7867 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 92.9757 | 85.1852 | monodelphis_domestica | ENSMODP00000002420 | 92.7389 | 84.9682 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 94.7637 | 90.0383 | bison_bison_bison | ENSBBBP00000011701 | 93.2161 | 88.5678 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 96.424 | 91.6986 | bison_bison_bison | ENSBBBP00000001944 | 96.424 | 91.6986 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 96.1686 | 92.2094 | monodon_monoceros | ENSMMNP00015013157 | 96.0459 | 92.0918 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 96.5517 | 92.7203 | sus_scrofa | ENSSSCP00000045253 | 96.4286 | 92.602 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 97.1903 | 94.636 | microcebus_murinus | ENSMICP00000008559 | 97.0663 | 94.5153 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 97.0626 | 92.2094 | octodon_degus | ENSODEP00000016482 | 96.9388 | 92.0918 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 96.424 | 91.5709 | jaculus_jaculus | ENSJJAP00000017805 | 96.301 | 91.4541 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 96.6794 | 91.954 | ovis_aries_rambouillet | ENSOARP00020000192 | 96.5561 | 91.8367 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 97.7011 | 94.3806 | ursus_maritimus | ENSUMAP00000025856 | 96.4691 | 93.1904 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 92.9757 | 89.3997 | ochotona_princeps | ENSOPRP00000009372 | 92.9757 | 89.3997 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 96.2963 | 92.3372 | delphinapterus_leucas | ENSDLEP00000025088 | 96.1735 | 92.2194 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 96.2963 | 92.4649 | physeter_catodon | ENSPCTP00005006846 | 96.1735 | 92.3469 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 96.5517 | 91.8263 | mus_caroli | MGP_CAROLIEiJ_P0025959 | 96.4286 | 91.7092 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 96.1686 | 91.1877 | mus_musculus | ENSMUSP00000020756 | 96.1686 | 91.1877 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 96.6794 | 91.6986 | dipodomys_ordii | ENSDORP00000010723 | 96.6794 | 91.6986 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 97.7011 | 93.8697 | mustela_putorius_furo | ENSMPUP00000002327 | 97.3282 | 93.5115 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 98.212 | 95.53 | aotus_nancymaae | ENSANAP00000011602 | 98.0867 | 95.4082 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 95.2746 | 93.1034 | saimiri_boliviensis_boliviensis | ENSSBOP00000032780 | 97.7719 | 95.5439 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 88.378 | 83.7803 | vicugna_pacos | ENSVPAP00000006606 | 88.378 | 83.7803 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 98.7229 | 97.5734 | chlorocebus_sabaeus | ENSCSAP00000010359 | 98.5969 | 97.449 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 67.3052 | 64.2401 | tupaia_belangeri | ENSTBEP00000009377 | 67.0483 | 63.9949 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 96.1686 | 91.4432 | mesocricetus_auratus | ENSMAUP00000013715 | 95.3165 | 90.6329 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 96.1686 | 92.3372 | phocoena_sinus | ENSPSNP00000016105 | 96.0459 | 92.2194 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 97.0626 | 92.7203 | camelus_dromedarius | ENSCDRP00005033003 | 91.018 | 86.9461 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 96.8072 | 93.742 | carlito_syrichta | ENSTSYP00000006076 | 96.6837 | 93.6225 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 97.7011 | 94.5083 | panthera_tigris_altaica | ENSPTIP00000015993 | 97.5765 | 94.3878 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 96.1686 | 91.6986 | rattus_norvegicus | ENSRNOP00000004919 | 96.0459 | 91.5816 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 21.7114 | 21.4559 | prolemur_simus | ENSPSMP00000020761 | 94.9721 | 93.8548 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 96.1686 | 91.8263 | microtus_ochrogaster | ENSMOCP00000003524 | 95.6798 | 91.3596 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 74.5849 | 71.1367 | sorex_araneus | ENSSARP00000011583 | 74.3003 | 70.8651 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 96.2963 | 91.3155 | peromyscus_maniculatus_bairdii | ENSPEMP00000003138 | 95.9288 | 90.9669 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 96.424 | 91.6986 | bos_mutus | ENSBMUP00000019285 | 96.424 | 91.6986 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 96.5517 | 91.4432 | mus_spicilegus | ENSMSIP00000020856 | 96.4286 | 91.3265 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 96.1686 | 91.4432 | cricetulus_griseus_chok1gshd | ENSCGRP00001010338 | 95.3165 | 90.6329 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 68.327 | 66.0281 | pteropus_vampyrus | ENSPVAP00000005095 | 68.2398 | 65.9439 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 96.5517 | 92.0817 | myotis_lucifugus | ENSMLUP00000003546 | 96.061 | 91.6137 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 92.9757 | 85.3129 | vombatus_ursinus | ENSVURP00010018257 | 92.7389 | 85.0955 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 97.0626 | 92.4649 | rhinolophus_ferrumequinum | ENSRFEP00010016004 | 96.9388 | 92.3469 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 97.4457 | 93.9974 | neovison_vison | ENSNVIP00000004471 | 97.4457 | 93.9974 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 96.424 | 91.6986 | bos_taurus | ENSBTAP00000011729 | 96.424 | 91.6986 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 18.2631 | 16.4751 | heterocephalus_glaber_female | ENSHGLP00000044389 | 79.8883 | 72.067 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 96.5517 | 91.5709 | heterocephalus_glaber_female | ENSHGLP00000035759 | 96.4286 | 91.4541 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 98.9783 | 97.4457 | rhinopithecus_bieti | ENSRBIP00000039096 | 98.9783 | 97.4457 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 97.5734 | 93.742 | canis_lupus_dingo | ENSCAFP00020034607 | 97.449 | 93.6225 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 97.0626 | 92.7203 | sciurus_vulgaris | ENSSVLP00005031411 | 97.0626 | 92.7203 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 93.2312 | 85.4406 | phascolarctos_cinereus | ENSPCIP00000012889 | 92.9936 | 85.2229 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 82.12 | 77.9055 | procavia_capensis | ENSPCAP00000010165 | 88.3242 | 83.7912 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 95.9132 | 91.4432 | nannospalax_galili | ENSNGAP00000021801 | 95.3046 | 90.8629 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 18.0077 | 16.986 | nannospalax_galili | ENSNGAP00000014346 | 89.2405 | 84.1772 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 96.424 | 91.6986 | bos_grunniens | ENSBGRP00000030096 | 96.424 | 91.6986 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 97.0626 | 92.4649 | chinchilla_lanigera | ENSCLAP00000008937 | 97.0626 | 92.4649 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 96.8072 | 92.0817 | cervus_hanglu_yarkandensis | ENSCHYP00000036836 | 96.6837 | 91.9643 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 96.8072 | 92.4649 | catagonus_wagneri | ENSCWAP00000015828 | 96.6837 | 92.3469 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 97.4457 | 92.848 | ictidomys_tridecemlineatus | ENSSTOP00000020583 | 97.3214 | 92.7296 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 96.1686 | 92.3372 | otolemur_garnettii | ENSOGAP00000008803 | 95.4373 | 91.635 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 97.5734 | 95.4023 | cebus_imitator | ENSCCAP00000040203 | 97.449 | 95.2806 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 97.4457 | 92.848 | urocitellus_parryii | ENSUPAP00010016090 | 97.3214 | 92.7296 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 96.424 | 91.5709 | moschus_moschiferus | ENSMMSP00000005141 | 94.4931 | 89.7372 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 98.9783 | 97.318 | rhinopithecus_roxellana | ENSRROP00000033947 | 98.9783 | 97.318 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 96.5517 | 91.6986 | mus_pahari | MGP_PahariEiJ_P0029993 | 96.4286 | 91.5816 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 80.4598 | 76.3729 | propithecus_coquereli | ENSPCOP00000008724 | 93.75 | 88.9881 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 55.6833 | 38.825 | caenorhabditis_elegans | BE0003N10.1.1 | 59.0786 | 41.1924 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 70.8812 | 54.4061 | drosophila_melanogaster | FBpp0290103 | 71.9844 | 55.2529 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 83.014 | 68.8378 | petromyzon_marinus | ENSPMAP00000009862 | 88.5559 | 73.4332 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 65.3895 | 51.341 | eptatretus_burgeri | ENSEBUP00000016450 | 77.8116 | 61.0942 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 88.8889 | 76.7561 | callorhinchus_milii | ENSCMIP00000005712 | 88.5496 | 76.4631 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 84.2912 | 73.0524 | latimeria_chalumnae | ENSLACP00000012552 | 91.1602 | 79.0055 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 87.9949 | 76.1175 | lepisosteus_oculatus | ENSLOCP00000018921 | 86.5578 | 74.8744 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 86.7178 | 71.6475 | carassius_auratus | ENSCARP00000063307 | 88.5267 | 73.1421 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 84.4189 | 72.9247 | astyanax_mexicanus | ENSAMXP00000034072 | 91.2983 | 78.8674 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 84.802 | 73.5632 | ictalurus_punctatus | ENSIPUP00000021638 | 93.1276 | 80.7854 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 89.0166 | 75.2235 | esox_lucius | ENSELUP00000003551 | 90.5195 | 76.4935 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 88.1226 | 74.7126 | electrophorus_electricus | ENSEEEP00000025438 | 89.7269 | 76.0728 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 89.7829 | 75.2235 | oncorhynchus_kisutch | ENSOKIP00005112008 | 91.2987 | 76.4935 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 86.9732 | 72.4138 | oncorhynchus_kisutch | ENSOKIP00005043258 | 86.2025 | 71.7721 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 89.6552 | 75.7344 | oncorhynchus_mykiss | ENSOMYP00000102299 | 90.1155 | 76.1232 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 89.7829 | 75.4789 | oncorhynchus_mykiss | ENSOMYP00000100027 | 91.2987 | 76.7533 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 89.6552 | 75.7344 | salmo_salar | ENSSSAP00000026318 | 90.1155 | 76.1232 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 88.2503 | 73.9464 | salmo_salar | ENSSSAP00000099626 | 88.9318 | 74.5174 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 88.8889 | 74.7126 | salmo_trutta | ENSSTUP00000043790 | 88.7755 | 74.6173 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 89.7829 | 75.6066 | salmo_trutta | ENSSTUP00000114016 | 90.2439 | 75.9949 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 87.9949 | 72.9247 | gadus_morhua | ENSGMOP00000034518 | 86.8852 | 72.005 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 88.5057 | 75.2235 | fundulus_heteroclitus | ENSFHEP00000024735 | 90 | 76.4935 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 84.2912 | 73.3078 | poecilia_reticulata | ENSPREP00000004419 | 90.6593 | 78.8462 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 88.1226 | 75.4789 | xiphophorus_maculatus | ENSXMAP00000016965 | 89.6104 | 76.7533 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 88.6335 | 75.2235 | oryzias_latipes | ENSORLP00000005430 | 87.1859 | 73.995 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 82.5032 | 69.4764 | cyclopterus_lumpus | ENSCLMP00005049052 | 83.1403 | 70.0129 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 86.7178 | 74.0741 | oreochromis_niloticus | ENSONIP00000001246 | 88.5267 | 75.6193 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 88.378 | 75.2235 | haplochromis_burtoni | ENSHBUP00000011309 | 89.8701 | 76.4935 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 88.5057 | 75.4789 | astatotilapia_calliptera | ENSACLP00000035876 | 90 | 76.7533 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 88.2503 | 74.4572 | sparus_aurata | ENSSAUP00010055481 | 89.7403 | 75.7143 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 88.8889 | 75.4789 | lates_calcarifer | ENSLCAP00010046933 | 90.3896 | 76.7533 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 89.0166 | 76.6283 | xenopus_tropicalis | ENSXETP00000013380 | 88.3397 | 76.0456 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 91.06 | 80.8429 | chrysemys_picta_bellii | ENSCPBP00000029593 | 89.4605 | 79.4228 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 89.9106 | 77.2669 | sphenodon_punctatus | ENSSPUP00000015250 | 90.2564 | 77.5641 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 87.1009 | 76.2452 | crocodylus_porosus | ENSCPRP00005019803 | 81.3842 | 71.2411 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 87.7395 | 75.8621 | laticauda_laticaudata | ENSLLTP00000023927 | 86.4151 | 74.717 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 77.3946 | 67.1775 | notechis_scutatus | ENSNSUP00000021505 | 84.4011 | 73.2591 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 81.8646 | 71.2644 | pseudonaja_textilis | ENSPTXP00000013449 | 79.2336 | 68.974 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 82.7586 | 72.9247 | anas_platyrhynchos_platyrhynchos | ENSAPLP00000021577 | 88.5246 | 78.0055 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 89.3997 | 78.5441 | gallus_gallus | ENSGALP00010027612 | 87.5 | 76.875 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 86.7178 | 76.2452 | meleagris_gallopavo | ENSMGAP00000001531 | 93.2692 | 82.0055 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 87.484 | 76.6283 | serinus_canaria | ENSSCAP00000019538 | 90.1316 | 78.9474 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 89.6552 | 78.5441 | parus_major | ENSPMJP00000019235 | 88.5246 | 77.5536 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 89.272 | 77.5223 | anolis_carolinensis | ENSACAP00000024053 | 88.5932 | 76.9328 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 69.3487 | 58.3653 | neolamprologus_brichardi | ENSNBRP00000016412 | 80.924 | 68.1073 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 88.1226 | 76.6283 | naja_naja | ENSNNAP00000003945 | 86.25 | 75 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 86.59 | 76.2452 | podarcis_muralis | ENSPMRP00000007094 | 88.7435 | 78.1414 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 87.484 | 76.6283 | geospiza_fortis | ENSGFOP00000005887 | 90.9695 | 79.6813 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 86.9732 | 76.6283 | taeniopygia_guttata | ENSTGUP00000007158 | 90.4382 | 79.6813 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 88.5057 | 75.7344 | erpetoichthys_calabaricus | ENSECRP00000032196 | 87.2796 | 74.6851 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 87.6117 | 74.3295 | kryptolebias_marmoratus | ENSKMAP00000023525 | 89.0909 | 75.5844 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 90.2937 | 79.8212 | salvator_merianae | ENSSMRP00000018936 | 88.2647 | 78.0275 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 87.9949 | 74.3295 | oryzias_javanicus | ENSOJAP00000022476 | 89.5969 | 75.6827 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 85.0575 | 72.9247 | amphilophus_citrinellus | ENSACIP00000013079 | 91.8621 | 78.7586 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 87.8672 | 73.5632 | dicentrarchus_labrax | ENSDLAP00005073222 | 89.2348 | 74.7082 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 85.0575 | 73.0524 | amphiprion_percula | ENSAPEP00000003263 | 91.358 | 78.4636 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 88.6335 | 75.2235 | oryzias_sinensis | ENSOSIP00000031183 | 87.1859 | 73.995 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 89.5275 | 78.9272 | pelodiscus_sinensis | ENSPSIP00000014870 | 91.395 | 80.5737 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 89.7829 | 80.5875 | gopherus_evgoodei | ENSGEVP00005004046 | 91.6558 | 82.2686 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 86.9732 | 78.6718 | chelonoidis_abingdonii | ENSCABP00000012290 | 88.5566 | 80.104 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 89.5275 | 75.3512 | seriola_dumerili | ENSSDUP00000025605 | 91.039 | 76.6234 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 88.2503 | 72.9247 | cottoperca_gobio | ENSCGOP00000052554 | 89.7403 | 74.1558 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 88.5057 | 78.1609 | struthio_camelus_australis | ENSSCUP00000009723 | 92.1543 | 81.383 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 81.0983 | 71.0089 | anser_brachyrhynchus | ENSABRP00000024886 | 83.4428 | 73.0618 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 87.7395 | 73.5632 | gasterosteus_aculeatus | ENSGACP00000014379 | 88.9896 | 74.6114 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 82.5032 | 69.4764 | cynoglossus_semilaevis | ENSCSEP00000001182 | 91.8919 | 77.3826 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 87.6117 | 75.6066 | poecilia_formosa | ENSPFOP00000010380 | 89.0909 | 76.8831 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 83.7803 | 71.1367 | myripristis_murdjan | ENSMMDP00005043225 | 87.9357 | 74.6649 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 88.7612 | 74.3295 | sander_lucioperca | ENSSLUP00000026998 | 90.4948 | 75.7812 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 89.272 | 78.4163 | strigops_habroptila | ENSSHBP00005003320 | 89.272 | 78.4163 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 88.7612 | 76.1175 | amphiprion_ocellaris | ENSAOCP00000029618 | 90.2597 | 77.4026 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 91.06 | 81.0983 | terrapene_carolina_triunguis | ENSTMTP00000019881 | 89.4605 | 79.6738 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 87.8672 | 74.2018 | hucho_hucho | ENSHHUP00000063113 | 90.8851 | 76.7503 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 89.9106 | 75.8621 | hucho_hucho | ENSHHUP00000028762 | 90.3723 | 76.2516 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 86.7178 | 73.4355 | betta_splendens | ENSBSLP00000053896 | 87.9534 | 74.4819 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 89.3997 | 77.7778 | pygocentrus_nattereri | ENSPNAP00000025638 | 91.0273 | 79.1938 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 88.378 | 75.4789 | anabas_testudineus | ENSATEP00000055442 | 89.8701 | 76.7533 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 89.3997 | 75.2235 | seriola_lalandi_dorsalis | ENSSLDP00000030396 | 90.9091 | 76.4935 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 88.7612 | 74.0741 | labrus_bergylta | ENSLBEP00000037098 | 90.4948 | 75.5208 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 85.0575 | 72.5415 | pundamilia_nyererei | ENSPNYP00000000477 | 88.5638 | 75.5319 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 87.7395 | 74.9681 | mastacembelus_armatus | ENSMAMP00000053718 | 89.4531 | 76.4323 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 87.7395 | 73.8186 | stegastes_partitus | ENSSPAP00000022417 | 89.2208 | 75.0649 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 88.6335 | 74.7126 | oncorhynchus_tshawytscha | ENSOTSP00005084639 | 88.6335 | 74.7126 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 89.5275 | 75.7344 | oncorhynchus_tshawytscha | ENSOTSP00005059246 | 47.047 | 39.7987 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 88.7612 | 75.7344 | acanthochromis_polyacanthus | ENSAPOP00000023575 | 90.2597 | 77.013 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 89.7829 | 79.3103 | aquila_chrysaetos_chrysaetos | ENSACCP00020007304 | 88.539 | 78.2116 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 87.7395 | 75.0958 | nothobranchius_furzeri | ENSNFUP00015006595 | 89.4531 | 76.5625 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 72.2861 | 61.3027 | cyprinodon_variegatus | ENSCVAP00000008198 | 90.56 | 76.8 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 88.8889 | 74.8404 | larimichthys_crocea | ENSLCRP00005020747 | 90.9804 | 76.6013 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 81.6092 | 67.6884 | takifugu_rubripes | ENSTRUP00000085824 | 80.1757 | 66.4994 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 87.484 | 76.5006 | ficedula_albicollis | ENSFALP00000011697 | 92.3181 | 80.7278 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 73.9464 | 60.5364 | hippocampus_comes | ENSHCOP00000001774 | 87.7273 | 71.8182 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 86.3346 | 71.3921 | cyprinus_carpio_carpio | ENSCCRP00000153998 | 86.0051 | 71.1196 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 79.1826 | 68.7101 | coturnix_japonica | ENSCJPP00005020937 | 88.8252 | 77.0774 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 87.7395 | 75.6066 | poecilia_latipinna | ENSPLAP00000006720 | 89.2208 | 76.8831 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 88.5057 | 75.4789 | maylandia_zebra | ENSMZEP00005036701 | 90 | 76.7533 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 85.5683 | 73.0524 | tetraodon_nigroviridis | ENSTNIP00000010268 | 89.6921 | 76.573 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 89.272 | 75.6066 | paramormyrops_kingsleyae | ENSPKIP00000004599 | 87.8141 | 74.3719 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 88.8889 | 77.5223 | scleropages_formosus | ENSSFOP00015072095 | 87.768 | 76.5448 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 87.2286 | 73.9464 | leptobrachium_leishanense | ENSLLEP00000027853 | 86.7853 | 73.5705 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 87.9949 | 73.9464 | scophthalmus_maximus | ENSSMAP00000030144 | 88.5604 | 74.4216 |
| ENSG00000138035 | homo_sapiens | ENSP00000400646 | 12.2605 | 10.8557 | oryzias_melastigma | ENSOMEP00000029022 | 71.6418 | 63.4328 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0000175 | enables 3-5-RNA exonuclease activity | 2 | IBA,IDA | Homo_sapiens(9606) | Function |
| GO:0000957 | involved_in mitochondrial RNA catabolic process | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0000958 | involved_in mitochondrial mRNA catabolic process | 2 | IBA,IDA | Homo_sapiens(9606) | Process |
| GO:0000962 | involved_in positive regulation of mitochondrial RNA catabolic process | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0000964 | involved_in mitochondrial RNA 5-end processing | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0000965 | involved_in mitochondrial RNA 3-end processing | 2 | IBA,IMP | Homo_sapiens(9606) | Process |
| GO:0003723 | enables RNA binding | 1 | HDA | Homo_sapiens(9606) | Function |
| GO:0004654 | enables polyribonucleotide nucleotidyltransferase activity | 2 | IBA,IDA | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005737 | located_in cytoplasm | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005739 | is_active_in mitochondrion | 2 | IBA,IDA | Homo_sapiens(9606) | Component |
| GO:0005758 | located_in mitochondrial intermembrane space | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005759 | located_in mitochondrial matrix | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0005789 | located_in endoplasmic reticulum membrane | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0005829 | is_active_in cytosol | 2 | IBA,IDA | Homo_sapiens(9606) | Component |
| GO:0006397 | involved_in mRNA processing | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0006401 | involved_in RNA catabolic process | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0006402 | involved_in mRNA catabolic process | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0008266 | enables poly(U) RNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0034046 | enables poly(G) binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0034599 | involved_in cellular response to oxidative stress | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0035198 | enables miRNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0035458 | involved_in cellular response to interferon-beta | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0035927 | involved_in RNA import into mitochondrion | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0035928 | involved_in rRNA import into mitochondrion | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0042788 | located_in polysomal ribosome | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0042802 | enables identical protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0043457 | involved_in regulation of cellular respiration | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0045025 | part_of mitochondrial degradosome | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0045926 | involved_in negative regulation of growth | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0051260 | involved_in protein homooligomerization | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0051591 | involved_in response to cAMP | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0051726 | involved_in regulation of cell cycle | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0060416 | involved_in response to growth hormone | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0061014 | involved_in positive regulation of mRNA catabolic process | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0070207 | involved_in protein homotrimerization | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0070584 | involved_in mitochondrion morphogenesis | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0071042 | involved_in nuclear polyadenylation-dependent mRNA catabolic process | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0097222 | involved_in mitochondrial mRNA polyadenylation | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0097421 | involved_in liver regeneration | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:2000627 | involved_in positive regulation of miRNA catabolic process | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:2000772 | involved_in regulation of cellular senescence | 1 | IDA | Homo_sapiens(9606) | Process |