Welcome to
RBP World!
Functions and Diseases
| RBP Type | Non-canonical_RBPs |
| Diseases | N.A. |
| Drug | Drug |
| Main interacting RNAs | N.A. |
| Moonlighting functions | Kinase |
| Localizations | N.A. |
| BulkPerturb-seq |
Description
| Ensembl ID | ENSG00000132155 | Gene ID | 5894 | Accession | 9829 |
| Symbol | RAF1 | Alias | NS5;CRAF;Raf-1;c-Raf;CMD1NN | Full Name | Raf-1 proto-oncogene, serine/threonine kinase |
| Status | Confidence | Length | 82101 bases | Strand | Minus strand |
| Position | 3 : 12582101 - 12664201 | RNA binding domain | N.A. | ||
| Summary | This gene is the cellular homolog of viral raf gene (v-raf). The encoded protein is a MAP kinase kinase kinase (MAP3K), which functions downstream of the Ras family of membrane associated GTPases to which it binds directly. Once activated, the cellular RAF1 protein can phosphorylate to activate the dual specificity protein kinases MEK1 and MEK2, which in turn phosphorylate to activate the serine/threonine specific protein kinases, ERK1 and ERK2. Activated ERKs are pleiotropic effectors of cell physiology and play an important role in the control of gene expression involved in the cell division cycle, apoptosis, cell differentiation and cell migration. Mutations in this gene are associated with Noonan syndrome 5 and LEOPARD syndrome 2. [provided by RefSeq, Jul 2008] | ||||
RNA binding domains (RBDs)
| Protein | Domain | Pfam ID | E-value | Domain number |
|---|
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|---|---|---|---|
| 20526349 | Rearrangements of the RAF kinase pathway in prostate cancer, gastric cancer and melanoma. | Nallasivam Palanisamy | 2010-07-01 | Nature medicine |
| 24556667 | Low false-positives in an mLumin-based bimolecular fluorescence complementation system with a bicistronic expression vector. | Shun Liu | 2014-02-19 | Sensors (Basel, Switzerland) |
| 26513016 | Lapatinib increases motility of triple-negative breast cancer cells by decreasing miRNA-7 and inducing Raf-1/MAPK-dependent interleukin-6. | Yu-Chun Hsiao | 2015-11-10 | Oncotarget |
| 16113238 | Site-specific biotinylation of RNA molecules by transcription using unnatural base pairs. | Kei Moriyama | 2005-08-19 | Nucleic acids research |
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| RAF1-236 | ENST00000688543 | 4662 | 615aa | ENSP00000509612 |
| RAF1-214 | ENST00000685348 | 4615 | 69aa | ENSP00000510285 |
| RAF1-253 | ENST00000690585 | 1586 | 91aa | ENSP00000510641 |
| RAF1-261 | ENST00000691888 | 1734 | 273aa | ENSP00000510730 |
| RAF1-201 | ENST00000251849 | 3191 | 648aa | ENSP00000251849 |
| RAF1-205 | ENST00000442415 | 3251 | 668aa | ENSP00000401888 |
| RAF1-235 | ENST00000688444 | 4972 | No protein | - |
| RAF1-257 | ENST00000691643 | 3906 | No protein | - |
| RAF1-218 | ENST00000685697 | 3588 | No protein | - |
| RAF1-264 | ENST00000692093 | 3187 | 615aa | ENSP00000509669 |
| RAF1-259 | ENST00000691724 | 3171 | 346aa | ENSP00000509255 |
| RAF1-203 | ENST00000423275 | 3034 | 69aa | ENSP00000401088 |
| RAF1-240 | ENST00000688803 | 4273 | No protein | - |
| RAF1-227 | ENST00000687326 | 5593 | 374aa | ENSP00000509665 |
| RAF1-206 | ENST00000460610 | 7152 | No protein | - |
| RAF1-249 | ENST00000689876 | 3198 | 494aa | ENSP00000508535 |
| RAF1-223 | ENST00000686455 | 5552 | No protein | - |
| RAF1-269 | ENST00000692830 | 2808 | 69aa | ENSP00000509461 |
| RAF1-213 | ENST00000684903 | 3129 | 69aa | ENSP00000508612 |
| RAF1-272 | ENST00000693312 | 2899 | 573aa | ENSP00000508686 |
| RAF1-263 | ENST00000692069 | 5746 | No protein | - |
| RAF1-237 | ENST00000688625 | 4704 | 69aa | ENSP00000509522 |
| RAF1-267 | ENST00000692773 | 3073 | 69aa | ENSP00000509055 |
| RAF1-252 | ENST00000690460 | 3141 | 644aa | ENSP00000509106 |
| RAF1-225 | ENST00000686762 | 3140 | 486aa | ENSP00000509767 |
| RAF1-273 | ENST00000693664 | 3079 | 499aa | ENSP00000509614 |
| RAF1-262 | ENST00000691899 | 2983 | 648aa | ENSP00000508763 |
| RAF1-243 | ENST00000689097 | 2877 | 69aa | ENSP00000509756 |
| RAF1-274 | ENST00000693705 | 2506 | 69aa | ENSP00000510697 |
| RAF1-233 | ENST00000688269 | 3413 | No protein | - |
| RAF1-231 | ENST00000687923 | 3031 | 611aa | ENSP00000510255 |
| RAF1-219 | ENST00000685738 | 3261 | 382aa | ENSP00000510156 |
| RAF1-251 | ENST00000690397 | 3037 | 611aa | ENSP00000508730 |
| RAF1-260 | ENST00000691779 | 2944 | 69aa | ENSP00000508592 |
| RAF1-266 | ENST00000692558 | 5400 | No protein | - |
| RAF1-265 | ENST00000692311 | 3641 | No protein | - |
| RAF1-250 | ENST00000689914 | 3215 | 374aa | ENSP00000509847 |
| RAF1-208 | ENST00000471449 | 1614 | No protein | - |
| RAF1-254 | ENST00000690625 | 3798 | No protein | - |
| RAF1-204 | ENST00000432427 | 2077 | 421aa | ENSP00000398591 |
| RAF1-245 | ENST00000689389 | 2899 | 589aa | ENSP00000510213 |
| RAF1-209 | ENST00000475353 | 5001 | No protein | - |
| RAF1-226 | ENST00000687257 | 5175 | No protein | - |
| RAF1-222 | ENST00000686409 | 6107 | No protein | - |
| RAF1-217 | ENST00000685653 | 4402 | 648aa | ENSP00000509968 |
| RAF1-248 | ENST00000689540 | 5062 | No protein | - |
| RAF1-256 | ENST00000691396 | 3104 | 80aa | ENSP00000510712 |
| RAF1-230 | ENST00000687505 | 2752 | No protein | - |
| RAF1-215 | ENST00000685437 | 2866 | 615aa | ENSP00000508794 |
| RAF1-246 | ENST00000689418 | 4974 | 69aa | ENSP00000509467 |
| RAF1-241 | ENST00000688914 | 1174 | No protein | - |
| RAF1-207 | ENST00000465826 | 1958 | No protein | - |
| RAF1-234 | ENST00000688326 | 1594 | 156aa | ENSP00000509907 |
| RAF1-212 | ENST00000494557 | 4000 | No protein | - |
| RAF1-232 | ENST00000687940 | 2650 | No protein | - |
| RAF1-247 | ENST00000689481 | 2102 | 69aa | ENSP00000510248 |
| RAF1-271 | ENST00000693069 | 1356 | 341aa | ENSP00000510072 |
| RAF1-210 | ENST00000491290 | 2084 | No protein | - |
| RAF1-229 | ENST00000687486 | 385 | 78aa | ENSP00000509469 |
| RAF1-224 | ENST00000686479 | 2234 | No protein | - |
| RAF1-255 | ENST00000691268 | 535 | 109aa | ENSP00000509128 |
| RAF1-216 | ENST00000685438 | 701 | No protein | - |
| RAF1-202 | ENST00000416093 | 801 | 87aa | ENSP00000391265 |
| RAF1-221 | ENST00000685959 | 963 | 278aa | ENSP00000510452 |
| RAF1-242 | ENST00000689033 | 1048 | 278aa | ENSP00000508983 |
| RAF1-268 | ENST00000692777 | 1946 | No protein | - |
| RAF1-244 | ENST00000689226 | 1215 | 287aa | ENSP00000510613 |
| RAF1-228 | ENST00000687348 | 774 | 152aa | ENSP00000509787 |
| RAF1-211 | ENST00000492690 | 633 | No protein | - |
| RAF1-238 | ENST00000688753 | 659 | 193aa | ENSP00000508771 |
| RAF1-220 | ENST00000685740 | 1098 | 193aa | ENSP00000510566 |
| RAF1-258 | ENST00000691718 | 1248 | 193aa | ENSP00000510160 |
| RAF1-270 | ENST00000692959 | 1429 | 193aa | ENSP00000509353 |
| RAF1-239 | ENST00000688779 | 568 | No protein | - |
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
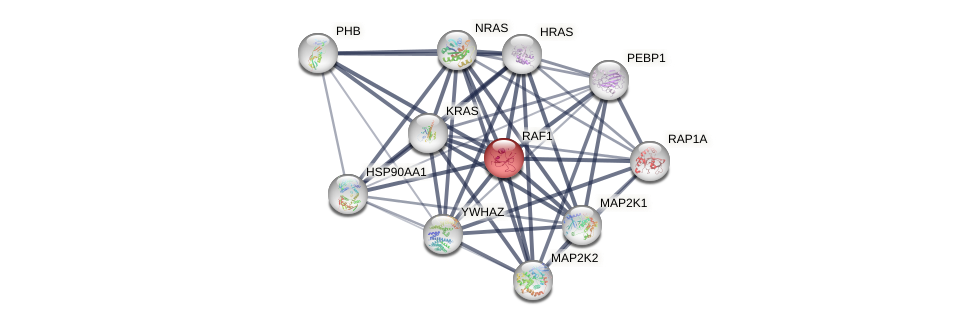
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0000165 | involved_in MAPK cascade | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0004672 | enables protein kinase activity | 1 | TAS | Homo_sapiens(9606) | Function |
| GO:0004674 | enables protein serine/threonine kinase activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0004709 | enables MAP kinase kinase kinase activity | 1 | IBA | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005524 | enables ATP binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0005634 | located_in nucleus | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0005737 | located_in cytoplasm | 2 | IDA,ISS | Homo_sapiens(9606) | Component |
| GO:0005739 | is_active_in mitochondrion | 1 | IBA | Homo_sapiens(9606) | Component |
| GO:0005741 | located_in mitochondrial outer membrane | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0005794 | located_in Golgi apparatus | 1 | IPI | Homo_sapiens(9606) | Component |
| GO:0005829 | is_active_in cytosol | 3 | IBA,IDA,TAS | Homo_sapiens(9606) | Component |
| GO:0005886 | colocalizes_with plasma membrane | 3 | IDA,TAS | Homo_sapiens(9606) | Component |
| GO:0006468 | involved_in protein phosphorylation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0006915 | acts_upstream_of_or_within_positive_effect apoptotic process | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0007165 | involved_in signal transduction | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0007190 | involved_in activation of adenylate cyclase activity | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0008285 | involved_in negative regulation of cell population proliferation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0008286 | involved_in insulin receptor signaling pathway | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0008625 | involved_in extrinsic apoptotic signaling pathway via death domain receptors | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0019899 | enables enzyme binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0030154 | involved_in cell differentiation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0030878 | involved_in thyroid gland development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0031143 | located_in pseudopodium | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0031267 | NOT enables small GTPase binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0031333 | involved_in negative regulation of protein-containing complex assembly | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0033138 | involved_in positive regulation of peptidyl-serine phosphorylation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0035019 | involved_in somatic stem cell population maintenance | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0035023 | involved_in regulation of Rho protein signal transduction | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0035773 | involved_in insulin secretion involved in cellular response to glucose stimulus | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0035994 | involved_in response to muscle stretch | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0042060 | involved_in wound healing | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0042802 | enables identical protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0042981 | involved_in regulation of apoptotic process | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0043066 | involved_in negative regulation of apoptotic process | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0043154 | involved_in negative regulation of cysteine-type endopeptidase activity involved in apoptotic process | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0043410 | involved_in positive regulation of MAPK cascade | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0044342 | involved_in type B pancreatic cell proliferation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0045104 | involved_in intermediate filament cytoskeleton organization | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0045595 | involved_in regulation of cell differentiation | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0045944 | involved_in positive regulation of transcription by RNA polymerase II | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0046872 | enables metal ion binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0048009 | involved_in insulin-like growth factor receptor signaling pathway | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0048011 | involved_in neurotrophin TRK receptor signaling pathway | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0048538 | involved_in thymus development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0060324 | involved_in face development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0071550 | involved_in death-inducing signaling complex assembly | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0106310 | enables protein serine kinase activity | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:1902042 | involved_in negative regulation of extrinsic apoptotic signaling pathway via death domain receptors | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:2000145 | involved_in regulation of cell motility | 1 | TAS | Homo_sapiens(9606) | Process |