Welcome to
RBP World!
Functions and Diseases
| RBP Type | Canonical_RBPs |
| Diseases | CancerSARS-COV-OC43FlavivirusesRVSARS-COV-2SINVDengueZika |
| Drug | N.A. |
| Main interacting RNAs | mRNA |
| Moonlighting functions | N.A. |
| Localizations | Stress granucle |
| BulkPerturb-seq | DataSet_01_148 DataSet_01_181 |
Description
| Ensembl ID | ENSG00000116001 | Gene ID | 7072 | Accession | 11802 |
| Symbol | TIA1 | Alias | WDM;ALS26;TIA-1 | Full Name | TIA1 cytotoxic granule associated RNA binding protein |
| Status | Confidence | Length | 39217 bases | Strand | Minus strand |
| Position | 2 : 70209444 - 70248660 | RNA binding domain | RRM_1 | ||
| Summary | The product encoded by this gene is a member of a RNA-binding protein family and possesses nucleolytic activity against cytotoxic lymphocyte (CTL) target cells. It has been suggested that this protein may be involved in the induction of apoptosis as it preferentially recognizes poly(A) homopolymers and induces DNA fragmentation in CTL targets. The major granule-associated species is a 15-kDa protein that is thought to be derived from the carboxyl terminus of the 40-kDa product by proteolytic processing. Alternative splicing resulting in different isoforms has been found for this gene. [provided by RefSeq, May 2017] | ||||
RNA binding domains (RBDs)
| Protein | Domain | Pfam ID | E-value | Domain number |
|---|---|---|---|---|
| ENSP00000404023 | RRM_1 | PF00076 | 9.7e-67 | 1 |
| ENSP00000404023 | RRM_1 | PF00076 | 9.7e-67 | 2 |
| ENSP00000404023 | RRM_1 | PF00076 | 9.7e-67 | 3 |
| ENSP00000282574 | RRM_1 | PF00076 | 1.1e-66 | 1 |
| ENSP00000282574 | RRM_1 | PF00076 | 1.1e-66 | 2 |
| ENSP00000282574 | RRM_1 | PF00076 | 1.1e-66 | 3 |
| ENSP00000401371 | RRM_1 | PF00076 | 1.1e-66 | 1 |
| ENSP00000401371 | RRM_1 | PF00076 | 1.1e-66 | 2 |
| ENSP00000401371 | RRM_1 | PF00076 | 1.1e-66 | 3 |
| ENSP00000399567 | RRM_1 | PF00076 | 1.1e-57 | 1 |
| ENSP00000399567 | RRM_1 | PF00076 | 1.1e-57 | 2 |
| ENSP00000399567 | RRM_1 | PF00076 | 1.1e-57 | 3 |
| ENSP00000413751 | RRM_1 | PF00076 | 1.4e-47 | 1 |
| ENSP00000413751 | RRM_1 | PF00076 | 1.4e-47 | 2 |
| ENSP00000354838 | RRM_1 | PF00076 | 1.9e-39 | 1 |
| ENSP00000354838 | RRM_1 | PF00076 | 1.9e-39 | 2 |
| ENSP00000430969 | RRM_1 | PF00076 | 1.4e-29 | 1 |
| ENSP00000430969 | RRM_1 | PF00076 | 1.4e-29 | 2 |
| ENSP00000402263 | RRM_1 | PF00076 | 8.1e-13 | 1 |
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|---|---|---|---|
| 35269506 | Dynamics of T-Cell Intracellular Antigen 1-Dependent Stress Granules in Proteostasis and Welander Distal Myopathy under Oxidative Stress. | Andrea Fernández-Gómez | 2022-03-04 | Cells |
| 33621982 | Tandem RNA binding sites induce self-association of the stress granule marker protein TIA-1. | E Fionna Loughlin | 2021-03-18 | Nucleic acids research |
| 23164372 | Regulated protein aggregation: stress granules and neurodegeneration. | Benjamin Wolozin | 2012-11-20 | Molecular neurodegeneration |
| 30828720 | The p53 mRNA: an integral part of the cellular stress response. | Lucia Haronikova | 2019-04-23 | Nucleic acids research |
| 22276125 | Role of the RNA-binding protein Nrd1 in stress granule formation and its implication in the stress response in fission yeast. | Ryosuke Satoh | 2012-01-01 | PloS one |
| 24824036 | The binding of TIA-1 to RNA C-rich sequences is driven by its C-terminal RRM domain. | Isabel Cruz-Gallardo | 2014-01-01 | RNA biology |
| 30327138 | Transcriptome meta-analysis identifies immune signature comprising of RNA binding proteins in ulcerative colitis patients. | Saima Naz | 2018-12-01 | Cellular immunology |
| 28221306 | Whole Exome Sequencing Identifies Atypical Welander Distal Myopathy in Patient. | Jennifer Gass | 2017-03-01 | Journal of clinical neuromuscular disease |
| 22875991 | Translation suppression promotes stress granule formation and cell survival in response to cold shock. | Sarah Hofmann | 2012-10-01 | Molecular biology of the cell |
| 32290247 | hnRNP A1 Regulates Alternative Splicing of Tau Exon 10 by Targeting 3' Splice Sites. | Yongchao Liu | 2020-04-10 | Cells |
| 29084250 | KSHV inhibits stress granule formation by viral ORF57 blocking PKR activation. | R Nishi Sharma | 2017-10-01 | PLoS pathogens |
| 16091628 | Novel DNA-binding properties of the RNA-binding protein TIAR. | A Esther Suswam | 2005-01-01 | Nucleic acids research |
| 31284549 | The Biological Axis of Protein Arginine Methylation and Asymmetric Dimethylarginine. | D Melody Fulton | 2019-07-06 | International journal of molecular sciences |
| 34345843 | Histone deacetylase inhibitors prevent H2O2 from inducing stress granule formation. | Siyuan Feng | 2020-06-10 | Current research in toxicology |
| 23902765 | RNA binding of T-cell intracellular antigen-1 (TIA-1) C-terminal RNA recognition motif is modified by pH conditions. | Isabel Cruz-Gallardo | 2013-09-06 | The Journal of biological chemistry |
| 28769895 | MicroRNAs as Important Players in Host-Adenovirus Interactions. | Diogo Piedade | 2017-01-01 | Frontiers in microbiology |
| 29429924 | The TIA1 RNA-Binding Protein Family Regulates EIF2AK2-Mediated Stress Response and Cell Cycle Progression. | Cindy Meyer | 2018-02-15 | Molecular cell |
| 23550649 | Control of alternative splicing in immune responses: many regulators, many predictions, much still to learn. | M Nicole Martinez | 2013-05-01 | Immunological reviews |
| 27271770 | Interleukin-1β induced Stress Granules Sequester COX-2 mRNA and Regulates its Stability and Translation in Human OA Chondrocytes. | Y Mohammad Ansari | 2016-06-08 | Scientific reports |
| 10938105 | The RNA-binding protein TIA-1 is a novel mammalian splicing regulator acting through intron sequences adjacent to a 5' splice site. | F F Del | 2000-09-01 | Molecular and cellular biology |
| 26824015 | Fas-activated Ser/Thr phosphoprotein (FAST) is a eukaryotic initiation factor 4E-binding protein that regulates mRNA stability and cell survival. | Wei Li | 2013-01-01 | Translation (Austin, Tex.) |
| 25200073 | Direct binding of the Alu binding protein dimer SRP9/14 to 40S ribosomal subunits promotes stress granule formation and is regulated by Alu RNA. | A Berger | 2014-01-01 | Nucleic acids research |
| 32605035 | The RNA Replication Site of Tula Orthohantavirus Resides within a Remodelled Golgi Network. | A Katherine Davies | 2020-06-27 | Cells |
| 10908586 | The drosophila melanogaster genome: translation factors and RNA binding proteins. | P Lasko | 2000-07-24 | The Journal of cell biology |
| 29504902 | TERIUS: accurate prediction of lncRNA via high-throughput sequencing data representing RNA-binding protein association. | Seo-Won Choi | 2018-02-19 | BMC bioinformatics |
| 31667556 | Mechanisms of secretion and spreading of pathological tau protein. | A Cecilia Brunello | 2020-05-01 | Cellular and molecular life sciences : CMLS |
| 27383630 | Combined structural, biochemical and cellular evidence demonstrates that both FGDF motifs in alphavirus nsP3 are required for efficient replication. | Tim Schulte | 2016-07-01 | Open biology |
| 24682828 | Structure, dynamics and RNA binding of the multi-domain splicing factor TIA-1. | Iren Wang | 2014-05-01 | Nucleic acids research |
| 25797890 | Active immunosurveillance in the tumor microenvironment of colorectal cancer is associated with low frequency tumor budding and improved outcome. | H Viktor Koelzer | 2015-08-01 | Translational research : the journal of laboratory and clinical medicine |
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| TIA1-205 | ENST00000433529 | 4634 | 386aa | ENSP00000401371 |
| TIA1-203 | ENST00000415783 | 4633 | 375aa | ENSP00000404023 |
| TIA1-201 | ENST00000282574 | 3823 | 385aa | ENSP00000282574 |
| TIA1-206 | ENST00000445587 | 1440 | 285aa | ENSP00000399567 |
| TIA1-216 | ENST00000486392 | 514 | No protein | - |
| TIA1-217 | ENST00000495774 | 533 | No protein | - |
| TIA1-207 | ENST00000454815 | 911 | 114aa | ENSP00000402263 |
| TIA1-208 | ENST00000468787 | 525 | No protein | - |
| TIA1-214 | ENST00000482876 | 562 | No protein | - |
| TIA1-220 | ENST00000497672 | 346 | No protein | - |
| TIA1-212 | ENST00000477415 | 510 | No protein | - |
| TIA1-218 | ENST00000496096 | 511 | No protein | - |
| TIA1-215 | ENST00000484065 | 350 | No protein | - |
| TIA1-204 | ENST00000416149 | 821 | 214aa | ENSP00000413751 |
| TIA1-209 | ENST00000474699 | 686 | No protein | - |
| TIA1-210 | ENST00000474809 | 915 | 144aa | ENSP00000430969 |
| TIA1-211 | ENST00000477044 | 904 | No protein | - |
| TIA1-202 | ENST00000361692 | 543 | 152aa | ENSP00000354838 |
| TIA1-219 | ENST00000496452 | 853 | No protein | - |
| TIA1-213 | ENST00000481650 | 549 | No protein | - |
Phenotypes
| ensgID | Trait | pValue | Pubmed ID |
|---|
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
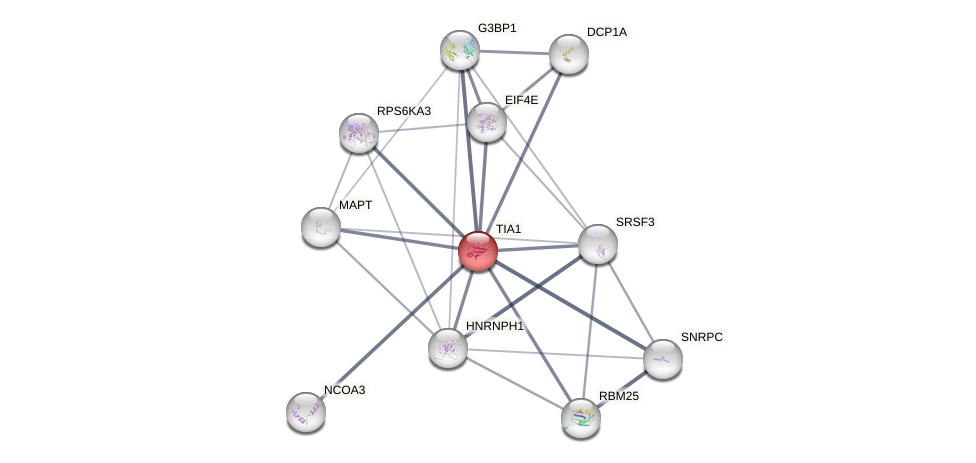
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000116001 | homo_sapiens | ENSP00000295971 | 22.9342 | 12.9848 | homo_sapiens | ENSP00000401371 | 35.2332 | 19.9482 |
| ENSG00000116001 | homo_sapiens | ENSP00000304139 | 21.9388 | 11.9898 | homo_sapiens | ENSP00000401371 | 22.2798 | 12.1762 |
| ENSG00000116001 | homo_sapiens | ENSP00000417451 | 7.49766 | 4.59231 | homo_sapiens | ENSP00000401371 | 20.7254 | 12.6943 |
| ENSG00000116001 | homo_sapiens | ENSP00000478691 | 32.8446 | 20.2346 | homo_sapiens | ENSP00000401371 | 29.0155 | 17.8756 |
| ENSG00000116001 | homo_sapiens | ENSP00000445366 | 26.0623 | 11.6147 | homo_sapiens | ENSP00000401371 | 23.8342 | 10.6218 |
| ENSG00000116001 | homo_sapiens | ENSP00000341826 | 31.7204 | 19.3548 | homo_sapiens | ENSP00000401371 | 30.5699 | 18.6528 |
| ENSG00000116001 | homo_sapiens | ENSP00000350090 | 36.875 | 22.8125 | homo_sapiens | ENSP00000401371 | 30.5699 | 18.9119 |
| ENSG00000116001 | homo_sapiens | ENSP00000493805 | 37.1875 | 22.5 | homo_sapiens | ENSP00000401371 | 30.829 | 18.6528 |
| ENSG00000116001 | homo_sapiens | ENSP00000376309 | 31.746 | 18.254 | homo_sapiens | ENSP00000401371 | 31.0881 | 17.8756 |
| ENSG00000116001 | homo_sapiens | ENSP00000284073 | 32.622 | 18.5976 | homo_sapiens | ENSP00000401371 | 27.7202 | 15.8031 |
| ENSG00000116001 | homo_sapiens | ENSP00000310471 | 30.3621 | 16.9916 | homo_sapiens | ENSP00000401371 | 28.2383 | 15.8031 |
| ENSG00000116001 | homo_sapiens | ENSP00000309166 | 29.1209 | 16.2088 | homo_sapiens | ENSP00000401371 | 27.4611 | 15.285 |
| ENSG00000116001 | homo_sapiens | ENSP00000498248 | 23.5897 | 13.8462 | homo_sapiens | ENSP00000401371 | 23.8342 | 13.9896 |
| ENSG00000116001 | homo_sapiens | ENSP00000359645 | 23.7852 | 14.0665 | homo_sapiens | ENSP00000401371 | 24.0933 | 14.2487 |
| ENSG00000116001 | homo_sapiens | ENSP00000257552 | 28.7293 | 16.8508 | homo_sapiens | ENSP00000401371 | 26.943 | 15.8031 |
| ENSG00000116001 | homo_sapiens | ENSP00000281722 | 25.8912 | 13.6961 | homo_sapiens | ENSP00000401371 | 35.7513 | 18.9119 |
| ENSG00000116001 | homo_sapiens | ENSP00000352956 | 21.6401 | 12.5285 | homo_sapiens | ENSP00000401371 | 24.6114 | 14.2487 |
| ENSG00000116001 | homo_sapiens | ENSP00000362936 | 46.3415 | 27.5261 | homo_sapiens | ENSP00000401371 | 34.456 | 20.4663 |
| ENSG00000116001 | homo_sapiens | ENSP00000313199 | 26.1972 | 15.2113 | homo_sapiens | ENSP00000401371 | 24.0933 | 13.9896 |
| ENSG00000116001 | homo_sapiens | ENSP00000253363 | 22.0755 | 12.4528 | homo_sapiens | ENSP00000401371 | 30.3109 | 17.0984 |
| ENSG00000116001 | homo_sapiens | ENSP00000363109 | 24.4027 | 13.1399 | homo_sapiens | ENSP00000401371 | 37.0466 | 19.9482 |
| ENSG00000116001 | homo_sapiens | ENSP00000316042 | 33.1148 | 20 | homo_sapiens | ENSP00000401371 | 26.1658 | 15.8031 |
| ENSG00000116001 | homo_sapiens | ENSP00000318195 | 14.507 | 6.76056 | homo_sapiens | ENSP00000401371 | 26.6839 | 12.4352 |
| ENSG00000116001 | homo_sapiens | ENSP00000394902 | 86.9333 | 79.2 | homo_sapiens | ENSP00000401371 | 84.456 | 76.943 |
| ENSG00000116001 | homo_sapiens | ENSP00000351108 | 29.2169 | 16.8675 | homo_sapiens | ENSP00000401371 | 25.1295 | 14.5078 |
| ENSG00000116001 | homo_sapiens | ENSP00000261741 | 11.6667 | 5.10417 | homo_sapiens | ENSP00000401371 | 29.0155 | 12.6943 |
| ENSG00000116001 | homo_sapiens | ENSP00000233078 | 17.9361 | 11.3022 | homo_sapiens | ENSP00000401371 | 18.9119 | 11.9171 |
| ENSG00000116001 | homo_sapiens | ENSP00000372127 | 17.1371 | 9.27419 | homo_sapiens | ENSP00000401371 | 22.0207 | 11.9171 |
| ENSG00000116001 | homo_sapiens | ENSP00000372105 | 17.1371 | 9.27419 | homo_sapiens | ENSP00000401371 | 22.0207 | 11.9171 |
| ENSG00000116001 | homo_sapiens | ENSP00000372484 | 17.1371 | 9.27419 | homo_sapiens | ENSP00000401371 | 22.0207 | 11.9171 |
| ENSG00000116001 | homo_sapiens | ENSP00000372154 | 17.1371 | 9.27419 | homo_sapiens | ENSP00000401371 | 22.0207 | 11.9171 |
| ENSG00000116001 | homo_sapiens | ENSP00000250831 | 17.3387 | 9.47581 | homo_sapiens | ENSP00000401371 | 22.2798 | 12.1762 |
| ENSG00000116001 | homo_sapiens | ENSP00000307155 | 17.3387 | 9.47581 | homo_sapiens | ENSP00000401371 | 22.2798 | 12.1762 |
| ENSG00000116001 | homo_sapiens | ENSP00000295470 | 23.3333 | 13.8095 | homo_sapiens | ENSP00000401371 | 25.3886 | 15.0259 |
| ENSG00000116001 | homo_sapiens | ENSP00000466025 | 23.2323 | 12.4579 | homo_sapiens | ENSP00000401371 | 17.8756 | 9.58549 |
| ENSG00000116001 | homo_sapiens | ENSP00000365950 | 36.9427 | 20.3822 | homo_sapiens | ENSP00000401371 | 15.0259 | 8.29016 |
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 93.2643 | 93.2643 | nomascus_leucogenys | ENSNLEP00000024597 | 100 | 100 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 100 | 100 | pan_paniscus | ENSPPAP00000036941 | 100 | 100 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 100 | 100 | pongo_abelii | ENSPPYP00000029885 | 100 | 100 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 96.3731 | 95.3368 | pan_troglodytes | ENSPTRP00000077003 | 96.3731 | 95.3368 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 91.1917 | 91.1917 | gorilla_gorilla | ENSGGOP00000051955 | 99.7167 | 99.7167 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 96.8912 | 93.5233 | ornithorhynchus_anatinus | ENSOANP00000037143 | 97.3958 | 94.0104 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 89.3782 | 88.0829 | sarcophilus_harrisii | ENSSHAP00000014697 | 95.5679 | 94.1828 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 74.3523 | 73.057 | notamacropus_eugenii | ENSMEUP00000012063 | 76.5333 | 75.2 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 81.6062 | 80.3109 | choloepus_hoffmanni | ENSCHOP00000003457 | 83.1135 | 81.7942 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 97.9275 | 97.4093 | dasypus_novemcinctus | ENSDNOP00000012684 | 89.3617 | 88.8889 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 73.3161 | 71.7617 | erinaceus_europaeus | ENSEEUP00000002724 | 72.7506 | 71.2082 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 99.7409 | 98.9637 | echinops_telfairi | ENSETEP00000008137 | 99.7409 | 98.9637 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 100 | 99.7409 | callithrix_jacchus | ENSCJAP00000087356 | 100 | 99.7409 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 100 | 99.7409 | cercocebus_atys | ENSCATP00000037522 | 100 | 99.7409 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 96.114 | 94.8186 | macaca_fascicularis | ENSMFAP00000054362 | 94.8849 | 93.6061 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 100 | 99.7409 | macaca_mulatta | ENSMMUP00000001495 | 100 | 99.7409 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 100 | 99.7409 | macaca_nemestrina | ENSMNEP00000012166 | 100 | 99.7409 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 34.1969 | 28.2383 | papio_anubis | ENSPANP00000005850 | 81.9876 | 67.7019 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 100 | 99.7409 | mandrillus_leucophaeus | ENSMLEP00000040877 | 100 | 99.7409 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 98.9637 | 98.1865 | tursiops_truncatus | ENSTTRP00000001119 | 98.9637 | 98.1865 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 93.0052 | 89.3782 | vulpes_vulpes | ENSVVUP00000039358 | 87.561 | 84.1463 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 98.7047 | 96.8912 | mus_spretus | MGP_SPRETEiJ_P0077704 | 98.7047 | 96.8912 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 99.2228 | 98.7047 | equus_asinus | ENSEASP00005010418 | 98.7113 | 98.1959 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 95.0777 | 93.5233 | capra_hircus | ENSCHIP00000022609 | 96.5789 | 95 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 100 | 98.9637 | loxodonta_africana | ENSLAFP00000002886 | 100 | 98.9637 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 83.6788 | 80.829 | panthera_leo | ENSPLOP00000016650 | 82.398 | 79.5918 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 99.4819 | 98.9637 | ailuropoda_melanoleuca | ENSAMEP00000016968 | 99.4819 | 98.9637 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 93.0052 | 90.6736 | bos_indicus_hybrid | ENSBIXP00005028241 | 87.7751 | 85.5746 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 99.4819 | 98.9637 | oryctolagus_cuniculus | ENSOCUP00000009639 | 99.4819 | 98.9637 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 99.2228 | 98.7047 | equus_caballus | ENSECAP00000059912 | 98.7113 | 98.1959 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 98.4456 | 97.4093 | canis_lupus_familiaris | ENSCAFP00845038068 | 96.2025 | 95.1899 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 99.4819 | 98.9637 | panthera_pardus | ENSPPRP00000021281 | 99.4819 | 98.9637 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 99.4819 | 98.7047 | marmota_marmota_marmota | ENSMMMP00000008719 | 99.4819 | 98.7047 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 99.7409 | 99.4819 | marmota_marmota_marmota | ENSMMMP00000005364 | 99.7409 | 99.4819 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 99.2228 | 98.4456 | balaenoptera_musculus | ENSBMSP00010022155 | 99.2228 | 98.4456 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 67.0984 | 65.0259 | balaenoptera_musculus | ENSBMSP00010028705 | 90.8772 | 88.0702 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 98.4456 | 97.6684 | cavia_porcellus | ENSCPOP00000028547 | 95.4774 | 94.7236 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 99.4819 | 98.9637 | felis_catus | ENSFCAP00000017232 | 99.4819 | 98.9637 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 99.4819 | 98.9637 | ursus_americanus | ENSUAMP00000033907 | 99.4819 | 98.9637 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 98.9637 | 97.6684 | monodelphis_domestica | ENSMODP00000028798 | 96.7089 | 95.443 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 98.9637 | 98.1865 | bison_bison_bison | ENSBBBP00000008929 | 98.9637 | 98.1865 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 85.4922 | 81.6062 | monodon_monoceros | ENSMMNP00015002688 | 91.4127 | 87.2576 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 99.2228 | 98.4456 | monodon_monoceros | ENSMMNP00015019884 | 99.2228 | 98.4456 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 99.4819 | 98.4456 | sus_scrofa | ENSSSCP00000026866 | 99.4819 | 98.4456 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 96.8912 | 95.0777 | microcebus_murinus | ENSMICP00000035736 | 96.3918 | 94.5876 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 100 | 99.4819 | octodon_degus | ENSODEP00000012708 | 100 | 99.4819 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 98.4456 | 97.4093 | jaculus_jaculus | ENSJJAP00000015985 | 98.4456 | 97.4093 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 98.9637 | 98.1865 | ovis_aries_rambouillet | ENSOARP00020007406 | 98.9637 | 98.1865 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 99.4819 | 98.9637 | ursus_maritimus | ENSUMAP00000029661 | 99.4819 | 98.9637 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 61.9171 | 61.1399 | ochotona_princeps | ENSOPRP00000000976 | 80.7432 | 79.7297 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 61.658 | 56.9948 | delphinapterus_leucas | ENSDLEP00000005146 | 84.0989 | 77.7385 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 50.2591 | 50 | delphinapterus_leucas | ENSDLEP00000007227 | 92.823 | 92.3445 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 63.2124 | 60.3627 | physeter_catodon | ENSPCTP00005025370 | 87.4552 | 83.5125 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 99.2228 | 98.4456 | physeter_catodon | ENSPCTP00005015009 | 99.2228 | 98.4456 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 98.4456 | 96.6321 | mus_caroli | MGP_CAROLIEiJ_P0074763 | 98.4456 | 96.6321 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 98.7047 | 96.6321 | mus_musculus | ENSMUSP00000093425 | 98.7047 | 96.6321 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 91.1917 | 90.1554 | dipodomys_ordii | ENSDORP00000008073 | 99.7167 | 98.5836 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 98.1865 | 97.1503 | mustela_putorius_furo | ENSMPUP00000007138 | 95.9494 | 94.9367 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 100 | 99.7409 | aotus_nancymaae | ENSANAP00000016518 | 100 | 99.7409 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 96.3731 | 95.0777 | saimiri_boliviensis_boliviensis | ENSSBOP00000036182 | 95.8763 | 94.5876 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 90.1554 | 89.3782 | vicugna_pacos | ENSVPAP00000006930 | 90.8616 | 90.0783 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 100 | 99.7409 | chlorocebus_sabaeus | ENSCSAP00000011235 | 100 | 99.7409 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 38.8601 | 38.601 | tupaia_belangeri | ENSTBEP00000008692 | 65.7895 | 65.3509 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 98.4456 | 97.1503 | mesocricetus_auratus | ENSMAUP00000023670 | 98.1912 | 96.8992 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 66.0622 | 61.658 | phocoena_sinus | ENSPSNP00000031032 | 83.3333 | 77.7778 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 96.3731 | 95.0777 | phocoena_sinus | ENSPSNP00000027229 | 99.2 | 97.8667 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 99.4819 | 98.7047 | camelus_dromedarius | ENSCDRP00005013911 | 99.4819 | 98.7047 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 98.1865 | 97.6684 | carlito_syrichta | ENSTSYP00000004833 | 97.9328 | 97.416 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 99.4819 | 98.9637 | panthera_tigris_altaica | ENSPTIP00000011667 | 99.4819 | 98.9637 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 97.4093 | 95.5959 | rattus_norvegicus | ENSRNOP00000094527 | 97.9167 | 96.0938 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 96.3731 | 94.8186 | prolemur_simus | ENSPSMP00000025337 | 95.8763 | 94.3299 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 97.4093 | 95.8549 | microtus_ochrogaster | ENSMOCP00000021619 | 97.1576 | 95.6072 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 75.3886 | 69.4301 | microtus_ochrogaster | ENSMOCP00000016301 | 84.104 | 77.4566 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 88.8601 | 88.342 | sorex_araneus | ENSSARP00000001814 | 99.4203 | 98.8406 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 98.7047 | 97.4093 | peromyscus_maniculatus_bairdii | ENSPEMP00000001411 | 98.7047 | 97.4093 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 98.9637 | 98.1865 | bos_mutus | ENSBMUP00000027806 | 98.9637 | 98.1865 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 98.7047 | 96.8912 | mus_spicilegus | ENSMSIP00000017885 | 98.7047 | 96.8912 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 98.1865 | 96.3731 | cricetulus_griseus_chok1gshd | ENSCGRP00001011898 | 97.9328 | 96.124 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 99.2228 | 98.7047 | pteropus_vampyrus | ENSPVAP00000004589 | 99.4805 | 98.961 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 97.9275 | 96.114 | myotis_lucifugus | ENSMLUP00000010625 | 97.4227 | 95.6186 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 98.9637 | 97.6684 | vombatus_ursinus | ENSVURP00010015505 | 96.7089 | 95.443 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 99.4819 | 98.9637 | rhinolophus_ferrumequinum | ENSRFEP00010022076 | 99.4819 | 98.9637 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 98.4456 | 97.4093 | neovison_vison | ENSNVIP00000004384 | 96.2025 | 95.1899 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 96.3731 | 93.7824 | bos_taurus | ENSBTAP00000070098 | 95.8763 | 93.299 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 99.7409 | 99.2228 | heterocephalus_glaber_female | ENSHGLP00000057631 | 92.7711 | 92.2892 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 83.1606 | 82.9016 | rhinopithecus_bieti | ENSRBIP00000013136 | 100 | 99.6885 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 96.6321 | 95.8549 | canis_lupus_dingo | ENSCAFP00020002119 | 99.4667 | 98.6667 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 99.7409 | 99.4819 | sciurus_vulgaris | ENSSVLP00005020400 | 99.7409 | 99.4819 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 98.9637 | 97.6684 | phascolarctos_cinereus | ENSPCIP00000017026 | 96.7089 | 95.443 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 81.8653 | 80.3109 | procavia_capensis | ENSPCAP00000001738 | 81.8653 | 80.3109 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 99.4819 | 98.4456 | nannospalax_galili | ENSNGAP00000009265 | 99.4819 | 98.4456 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 96.114 | 95.0777 | bos_grunniens | ENSBGRP00000008561 | 90.9314 | 89.951 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 99.7409 | 99.2228 | chinchilla_lanigera | ENSCLAP00000014297 | 99.7409 | 99.2228 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 93.2643 | 90.6736 | cervus_hanglu_yarkandensis | ENSCHYP00000019141 | 88.0196 | 85.5746 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 99.4819 | 98.4456 | catagonus_wagneri | ENSCWAP00000004982 | 99.4819 | 98.4456 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 98.7047 | 97.6684 | ictidomys_tridecemlineatus | ENSSTOP00000007449 | 97.9434 | 96.9152 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 93.0052 | 90.9326 | otolemur_garnettii | ENSOGAP00000009465 | 91.8159 | 89.7698 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 91.4508 | 90.9326 | cebus_imitator | ENSCCAP00000037118 | 100 | 99.4334 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 99.7409 | 99.4819 | urocitellus_parryii | ENSUPAP00010006958 | 99.7409 | 99.4819 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 98.9637 | 98.1865 | moschus_moschiferus | ENSMMSP00000013345 | 98.9637 | 98.1865 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 100 | 99.7409 | rhinopithecus_roxellana | ENSRROP00000017266 | 100 | 99.7409 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 98.4456 | 96.3731 | mus_pahari | MGP_PahariEiJ_P0050871 | 98.4456 | 96.3731 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 82.9016 | 82.6425 | propithecus_coquereli | ENSPCOP00000004934 | 100 | 99.6875 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 92.228 | 88.601 | latimeria_chalumnae | ENSLACP00000020283 | 89.6725 | 86.1461 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 91.4508 | 85.7513 | lepisosteus_oculatus | ENSLOCP00000018857 | 92.8947 | 87.1053 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 56.2176 | 50.5181 | clupea_harengus | ENSCHAP00000048935 | 80.6691 | 72.4907 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 90.6736 | 82.6425 | danio_rerio | ENSDARP00000068889 | 90.6736 | 82.6425 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 88.0829 | 80.5699 | carassius_auratus | ENSCARP00000020840 | 87.8553 | 80.3618 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 89.6373 | 81.0881 | carassius_auratus | ENSCARP00000010848 | 89.6373 | 81.0881 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 88.342 | 80.829 | esox_lucius | ENSELUP00000053888 | 85.6784 | 78.392 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 87.3057 | 80.0518 | esox_lucius | ENSELUP00000075433 | 85.101 | 78.0303 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 88.342 | 81.3472 | oncorhynchus_kisutch | ENSOKIP00005093743 | 86.7684 | 79.8982 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 88.8601 | 81.3472 | oncorhynchus_kisutch | ENSOKIP00005066409 | 86.8354 | 79.4937 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 86.2694 | 78.2383 | oncorhynchus_kisutch | ENSOKIP00005010507 | 84.7328 | 76.8448 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 86.2694 | 79.5337 | oncorhynchus_kisutch | ENSOKIP00005007096 | 81.2195 | 74.8781 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 89.1192 | 81.3472 | oncorhynchus_mykiss | ENSOMYP00000001840 | 87.9795 | 80.3069 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 88.601 | 81.6062 | oncorhynchus_mykiss | ENSOMYP00000065264 | 87.2449 | 80.3571 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 86.2694 | 79.5337 | oncorhynchus_mykiss | ENSOMYP00000011490 | 81.2195 | 74.8781 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 87.8238 | 79.2746 | oncorhynchus_mykiss | ENSOMYP00000082789 | 83.9109 | 75.7426 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 87.0466 | 79.7927 | salmo_salar | ENSSSAP00000116470 | 83.1683 | 76.2376 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 87.5648 | 79.0155 | salmo_salar | ENSSSAP00000110742 | 88.7139 | 80.0525 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 88.601 | 81.3472 | salmo_salar | ENSSSAP00000104626 | 85.5 | 78.5 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 90.4145 | 82.9016 | salmo_salar | ENSSSAP00000104620 | 89.4872 | 82.0513 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 89.1192 | 81.3472 | salmo_trutta | ENSSTUP00000095176 | 87.9795 | 80.3069 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 88.601 | 81.3472 | salmo_trutta | ENSSTUP00000014800 | 85.5 | 78.5 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 88.601 | 80.829 | salmo_trutta | ENSSTUP00000003074 | 79.7203 | 72.7273 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 88.8601 | 81.6062 | salmo_trutta | ENSSTUP00000048673 | 87.7238 | 80.5627 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 86.0104 | 75.3886 | gadus_morhua | ENSGMOP00000009018 | 85.1282 | 74.6154 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 87.0466 | 77.7202 | gadus_morhua | ENSGMOP00000066689 | 80.1909 | 71.599 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 87.3057 | 77.9793 | fundulus_heteroclitus | ENSFHEP00000002026 | 87.3057 | 77.9793 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 89.1192 | 80.829 | fundulus_heteroclitus | ENSFHEP00000005178 | 88.8889 | 80.6202 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 88.8601 | 80.3109 | poecilia_reticulata | ENSPREP00000025709 | 81.8616 | 73.9857 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 87.5648 | 78.2383 | poecilia_reticulata | ENSPREP00000028428 | 87.5648 | 78.2383 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 88.8601 | 80.829 | xiphophorus_maculatus | ENSXMAP00000021522 | 88.8601 | 80.829 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 86.5285 | 77.2021 | xiphophorus_maculatus | ENSXMAP00000039580 | 88.8298 | 79.2553 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 88.342 | 79.0155 | oryzias_latipes | ENSORLP00000003146 | 88.342 | 79.0155 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 89.1192 | 81.3472 | oryzias_latipes | ENSORLP00000019581 | 89.1192 | 81.3472 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 87.3057 | 77.9793 | cyclopterus_lumpus | ENSCLMP00005021739 | 86.8557 | 77.5773 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 86.5285 | 78.2383 | cyclopterus_lumpus | ENSCLMP00005037628 | 81.8627 | 74.0196 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 89.8964 | 81.0881 | oreochromis_niloticus | ENSONIP00000034900 | 85.468 | 77.0936 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 83.1606 | 74.3523 | oreochromis_niloticus | ENSONIP00000027286 | 82.0972 | 73.4015 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 89.8964 | 81.6062 | haplochromis_burtoni | ENSHBUP00000020871 | 89.6641 | 81.3953 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 87.3057 | 78.2383 | haplochromis_burtoni | ENSHBUP00000024935 | 87.3057 | 78.2383 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 89.8964 | 81.6062 | astatotilapia_calliptera | ENSACLP00000003417 | 89.6641 | 81.3953 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 65.8031 | 60.1036 | astatotilapia_calliptera | ENSACLP00000039235 | 91.3669 | 83.4532 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 89.3782 | 81.8653 | sparus_aurata | ENSSAUP00010053210 | 89.3782 | 81.8653 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 86.2694 | 77.7202 | sparus_aurata | ENSSAUP00010032250 | 83.0424 | 74.813 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 87.8238 | 79.0155 | lates_calcarifer | ENSLCAP00010016892 | 88.0519 | 79.2208 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 90.1554 | 82.3834 | lates_calcarifer | ENSLCAP00010033053 | 89.9225 | 82.1705 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 94.8186 | 86.7876 | xenopus_tropicalis | ENSXETP00000097567 | 92.6582 | 84.8101 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 97.6684 | 95.5959 | chrysemys_picta_bellii | ENSCPBP00000004475 | 98.1771 | 96.0938 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 97.9275 | 94.3005 | sphenodon_punctatus | ENSSPUP00000011118 | 98.4375 | 94.7917 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 97.6684 | 95.0777 | crocodylus_porosus | ENSCPRP00005023613 | 98.1771 | 95.5729 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 95.3368 | 92.228 | laticauda_laticaudata | ENSLLTP00000006949 | 94.6015 | 91.5167 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 97.1503 | 94.0415 | notechis_scutatus | ENSNSUP00000028594 | 95.6633 | 92.602 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 97.1503 | 94.3005 | pseudonaja_textilis | ENSPTXP00000023898 | 95.6633 | 92.8571 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 97.6684 | 95.3368 | anas_platyrhynchos_platyrhynchos | ENSAPLP00000009697 | 98.4334 | 96.0835 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 93.5233 | 90.4145 | gallus_gallus | ENSGALP00010026686 | 84.5433 | 81.733 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 87.8238 | 83.9378 | meleagris_gallopavo | ENSMGAP00000003240 | 94.958 | 90.7563 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 85.2332 | 80.0518 | serinus_canaria | ENSSCAP00000020764 | 91.1357 | 85.5956 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 40.9326 | 39.3782 | parus_major | ENSPMJP00000021316 | 96.9325 | 93.2515 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 16.0622 | 13.9896 | parus_major | ENSPMJP00000021317 | 77.5 | 67.5 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 66.5803 | 61.1399 | neolamprologus_brichardi | ENSNBRP00000013098 | 92.7798 | 85.1986 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 87.3057 | 78.2383 | neolamprologus_brichardi | ENSNBRP00000014660 | 87.3057 | 78.2383 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 91.1917 | 88.0829 | naja_naja | ENSNNAP00000007918 | 88 | 85 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 88.0829 | 85.4922 | podarcis_muralis | ENSPMRP00000018385 | 95.2381 | 92.437 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 80.829 | 75.1295 | geospiza_fortis | ENSGFOP00000018854 | 85.9504 | 79.8898 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 61.1399 | 56.9948 | taeniopygia_guttata | ENSTGUP00000034180 | 85.5072 | 79.7101 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 88.601 | 84.456 | taeniopygia_guttata | ENSTGUP00000017586 | 93.956 | 89.5604 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 86.0104 | 81.3472 | erpetoichthys_calabaricus | ENSECRP00000000404 | 87.5989 | 82.8496 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 87.8238 | 79.0155 | kryptolebias_marmoratus | ENSKMAP00000015561 | 87.8238 | 79.0155 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 89.1192 | 81.6062 | kryptolebias_marmoratus | ENSKMAP00000010917 | 89.1192 | 81.6062 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 97.1503 | 94.3005 | salvator_merianae | ENSSMRP00000019927 | 95.6633 | 92.8571 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 76.943 | 68.9119 | oryzias_javanicus | ENSOJAP00000040801 | 80.0539 | 71.6981 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 89.1192 | 81.3472 | oryzias_javanicus | ENSOJAP00000012948 | 88.8889 | 81.1369 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 83.6788 | 77.2021 | amphilophus_citrinellus | ENSACIP00000018433 | 92.2857 | 85.1429 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 85.7513 | 77.4611 | amphilophus_citrinellus | ENSACIP00000021067 | 89.9457 | 81.25 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 85.4922 | 77.7202 | dicentrarchus_labrax | ENSDLAP00005014141 | 81.4815 | 74.0741 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 87.0466 | 78.7565 | dicentrarchus_labrax | ENSDLAP00005077573 | 85.2792 | 77.1574 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 86.5285 | 79.5337 | amphiprion_percula | ENSAPEP00000003513 | 91.7582 | 84.3407 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 88.0829 | 78.4974 | amphiprion_percula | ENSAPEP00000004470 | 88.0829 | 78.4974 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 74.6114 | 64.5078 | oryzias_sinensis | ENSOSIP00000035686 | 81.3559 | 70.339 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 89.1192 | 81.3472 | oryzias_sinensis | ENSOSIP00000048955 | 89.1192 | 81.3472 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 13.7306 | 11.399 | oryzias_sinensis | ENSOSIP00000022978 | 81.5385 | 67.6923 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 13.7306 | 11.399 | oryzias_sinensis | ENSOSIP00000022975 | 81.5385 | 67.6923 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 93.7824 | 90.9326 | pelodiscus_sinensis | ENSPSIP00000015263 | 97.0509 | 94.1019 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 97.6684 | 95.5959 | gopherus_evgoodei | ENSGEVP00005008885 | 98.1771 | 96.0938 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 92.487 | 90.1554 | chelonoidis_abingdonii | ENSCABP00000029980 | 93.4555 | 91.0995 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 88.0829 | 79.2746 | seriola_dumerili | ENSSDUP00000032760 | 88.3117 | 79.4805 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 90.1554 | 82.6425 | seriola_dumerili | ENSSDUP00000004668 | 89.9225 | 82.4289 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 89.8964 | 82.3834 | cottoperca_gobio | ENSCGOP00000041406 | 89.433 | 81.9588 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 87.8238 | 79.0155 | cottoperca_gobio | ENSCGOP00000043918 | 87.5969 | 78.8114 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 88.0829 | 86.0104 | struthio_camelus_australis | ENSSCUP00000002914 | 98.2659 | 95.9538 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 97.6684 | 95.3368 | anser_brachyrhynchus | ENSABRP00000023545 | 97.9221 | 95.5844 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 78.7565 | 72.0207 | gasterosteus_aculeatus | ENSGACP00000017578 | 93.8272 | 85.8025 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 87.3057 | 77.7202 | gasterosteus_aculeatus | ENSGACP00000020847 | 86.6324 | 77.1208 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 87.0466 | 76.6839 | cynoglossus_semilaevis | ENSCSEP00000022494 | 87.2727 | 76.8831 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 87.5648 | 79.5337 | cynoglossus_semilaevis | ENSCSEP00000022242 | 87.3385 | 79.3282 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 85.7513 | 77.9793 | cynoglossus_semilaevis | ENSCSEP00000015594 | 81.7284 | 74.321 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 86.7876 | 77.7202 | poecilia_formosa | ENSPFOP00000027004 | 86.7876 | 77.7202 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 88.342 | 80.3109 | poecilia_formosa | ENSPFOP00000004497 | 88.1137 | 80.1034 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 82.1244 | 75.1295 | myripristis_murdjan | ENSMMDP00005052212 | 63.7827 | 58.3501 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 87.8238 | 79.0155 | sander_lucioperca | ENSSLUP00000024707 | 87.5969 | 78.8114 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 88.8601 | 81.6062 | sander_lucioperca | ENSSLUP00000043028 | 83.6585 | 76.8293 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 96.6321 | 92.7461 | strigops_habroptila | ENSSHBP00005007140 | 97.389 | 93.4726 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 88.0829 | 78.4974 | amphiprion_ocellaris | ENSAOCP00000014114 | 88.3117 | 78.7013 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 89.3782 | 81.3472 | amphiprion_ocellaris | ENSAOCP00000007363 | 89.1473 | 81.1369 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 85.4922 | 83.1606 | terrapene_carolina_triunguis | ENSTMTP00000006948 | 98.2143 | 95.5357 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 89.6373 | 81.0881 | hucho_hucho | ENSHHUP00000055130 | 88.2653 | 79.8469 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 90.4145 | 82.9016 | hucho_hucho | ENSHHUP00000005836 | 89.4872 | 82.0513 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 90.4145 | 83.4197 | hucho_hucho | ENSHHUP00000036635 | 89.4872 | 82.5641 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 88.8601 | 81.3472 | hucho_hucho | ENSHHUP00000085009 | 87.7238 | 80.3069 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 87.8238 | 78.2383 | betta_splendens | ENSBSLP00000009950 | 88.2812 | 78.6458 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 90.1554 | 81.8653 | betta_splendens | ENSBSLP00000016108 | 89.9225 | 81.6537 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 86.0104 | 77.4611 | anabas_testudineus | ENSATEP00000042699 | 86.2338 | 77.6623 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 89.8964 | 81.6062 | anabas_testudineus | ENSATEP00000057634 | 89.6641 | 81.3953 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 88.0829 | 79.2746 | seriola_lalandi_dorsalis | ENSSLDP00000014341 | 88.3117 | 79.4805 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 90.1554 | 82.6425 | seriola_lalandi_dorsalis | ENSSLDP00000018608 | 89.9225 | 82.4289 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 89.8964 | 81.8653 | labrus_bergylta | ENSLBEP00000000164 | 89.6641 | 81.6537 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 87.3057 | 77.4611 | labrus_bergylta | ENSLBEP00000030604 | 87.5325 | 77.6623 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 87.3057 | 78.2383 | pundamilia_nyererei | ENSPNYP00000008655 | 87.3057 | 78.2383 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 89.8964 | 81.6062 | pundamilia_nyererei | ENSPNYP00000011058 | 89.6641 | 81.3953 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 89.6373 | 82.1244 | mastacembelus_armatus | ENSMAMP00000048397 | 89.4057 | 81.9121 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 87.8238 | 78.2383 | mastacembelus_armatus | ENSMAMP00000005476 | 88.0519 | 78.4416 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 87.5648 | 78.2383 | stegastes_partitus | ENSSPAP00000010844 | 87.5648 | 78.2383 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 24.8705 | 19.4301 | stegastes_partitus | ENSSPAP00000002990 | 67.6056 | 52.8169 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 84.456 | 75.9067 | stegastes_partitus | ENSSPAP00000005858 | 89.5604 | 80.4945 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 85.4922 | 76.1658 | oncorhynchus_tshawytscha | ENSOTSP00005068542 | 75.5149 | 67.2769 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 88.8601 | 81.6062 | oncorhynchus_tshawytscha | ENSOTSP00005088743 | 87.7238 | 80.5627 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 88.8601 | 81.6062 | oncorhynchus_tshawytscha | ENSOTSP00005026333 | 87.7238 | 80.5627 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 85.2332 | 76.1658 | oncorhynchus_tshawytscha | ENSOTSP00005097477 | 83.715 | 74.8092 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 85.2332 | 76.1658 | oncorhynchus_tshawytscha | ENSOTSP00005097397 | 83.715 | 74.8092 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 88.8601 | 81.8653 | oncorhynchus_tshawytscha | ENSOTSP00005027513 | 78.6697 | 72.4771 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 89.3782 | 81.3472 | acanthochromis_polyacanthus | ENSAPOP00000000665 | 89.3782 | 81.3472 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 83.6788 | 75.3886 | acanthochromis_polyacanthus | ENSAPOP00000024459 | 82.1883 | 74.0458 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 97.1503 | 94.8186 | aquila_chrysaetos_chrysaetos | ENSACCP00020013206 | 97.9112 | 95.5614 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 87.5648 | 78.4974 | nothobranchius_furzeri | ENSNFUP00015010846 | 87.5648 | 78.4974 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 89.3782 | 81.0881 | nothobranchius_furzeri | ENSNFUP00015051290 | 88.9175 | 80.6701 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 40.9326 | 37.5648 | cyprinodon_variegatus | ENSCVAP00000023004 | 92.9412 | 85.2941 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 81.8653 | 73.8342 | cyprinodon_variegatus | ENSCVAP00000011341 | 86.8132 | 78.2967 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 88.601 | 80.5699 | cyprinodon_variegatus | ENSCVAP00000028425 | 88.3721 | 80.3618 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 15.544 | 13.2124 | cyprinodon_variegatus | ENSCVAP00000023010 | 88.2353 | 75 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 89.1192 | 81.0881 | denticeps_clupeoides | ENSDCDP00000039455 | 82.2967 | 74.8804 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 89.8964 | 82.3834 | larimichthys_crocea | ENSLCRP00005063147 | 89.6641 | 82.1705 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 87.0466 | 78.7565 | larimichthys_crocea | ENSLCRP00005034654 | 85.7143 | 77.551 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 87.5648 | 79.7927 | takifugu_rubripes | ENSTRUP00000081424 | 85.7868 | 78.1726 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 86.5285 | 78.2383 | takifugu_rubripes | ENSTRUP00000062103 | 86.5285 | 78.2383 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 54.9223 | 52.8497 | ficedula_albicollis | ENSFALP00000008688 | 95.9276 | 92.3077 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 38.8601 | 29.7927 | ficedula_albicollis | ENSFALP00000026967 | 36.5854 | 28.0488 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 87.0466 | 77.9793 | hippocampus_comes | ENSHCOP00000004194 | 88.6544 | 79.4195 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 88.8601 | 81.6062 | hippocampus_comes | ENSHCOP00000003963 | 88.6305 | 81.3953 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 85.4922 | 78.2383 | cyprinus_carpio_carpio | ENSCCRP00000020533 | 82.9146 | 75.8794 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 84.456 | 75.1295 | cyprinus_carpio_carpio | ENSCCRP00000020535 | 81.0945 | 72.1393 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 97.4093 | 94.8186 | coturnix_japonica | ENSCJPP00005013985 | 98.1723 | 95.5614 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 85.2332 | 77.9793 | poecilia_latipinna | ENSPLAP00000025870 | 91.3889 | 83.6111 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 86.7876 | 77.7202 | poecilia_latipinna | ENSPLAP00000007098 | 86.7876 | 77.7202 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 71.7617 | 62.4352 | sinocyclocheilus_grahami | ENSSGRP00000008974 | 74.0642 | 64.4385 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 89.8964 | 81.6062 | maylandia_zebra | ENSMZEP00005002291 | 89.6641 | 81.3953 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 87.3057 | 78.2383 | maylandia_zebra | ENSMZEP00005007899 | 87.3057 | 78.2383 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 89.1192 | 81.6062 | tetraodon_nigroviridis | ENSTNIP00000014378 | 87.9795 | 80.5627 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 88.8601 | 79.7927 | tetraodon_nigroviridis | ENSTNIP00000016465 | 87.7238 | 78.7724 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 24.8705 | 23.3161 | paramormyrops_kingsleyae | ENSPKIP00000003089 | 96 | 90 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 89.8964 | 84.9741 | paramormyrops_kingsleyae | ENSPKIP00000039864 | 89.8964 | 84.9741 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 87.3057 | 81.6062 | scleropages_formosus | ENSSFOP00015028824 | 86.6324 | 80.9769 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 89.1192 | 83.6788 | scleropages_formosus | ENSSFOP00015056109 | 80.3738 | 75.4673 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 94.0415 | 87.8238 | leptobrachium_leishanense | ENSLLEP00000007857 | 82.6879 | 77.221 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 89.6373 | 81.8653 | scophthalmus_maximus | ENSSMAP00000033016 | 89.4057 | 81.6537 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 86.7876 | 77.2021 | scophthalmus_maximus | ENSSMAP00000050683 | 86.5633 | 77.0026 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 88.342 | 79.2746 | oryzias_melastigma | ENSOMEP00000014937 | 82.9684 | 74.4526 |
| ENSG00000116001 | homo_sapiens | ENSP00000401371 | 89.1192 | 81.3472 | oryzias_melastigma | ENSOMEP00000019116 | 89.1192 | 81.3472 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0000381 | involved_in regulation of alternative mRNA splicing, via spliceosome | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0001818 | involved_in negative regulation of cytokine production | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0003723 | enables RNA binding | 2 | HDA,IDA | Homo_sapiens(9606) | Function |
| GO:0003730 | enables mRNA 3-UTR binding | 1 | ISS | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005634 | located_in nucleus | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005654 | located_in nucleoplasm | 2 | IDA,TAS | Homo_sapiens(9606) | Component |
| GO:0005737 | located_in cytoplasm | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005829 | located_in cytosol | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0006397 | involved_in mRNA processing | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0006915 | involved_in apoptotic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0008143 | enables poly(A) binding | 1 | TAS | Homo_sapiens(9606) | Function |
| GO:0008380 | involved_in RNA splicing | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0010494 | located_in cytoplasmic stress granule | 2 | IDA,ISS | Homo_sapiens(9606) | Component |
| GO:0017148 | involved_in negative regulation of translation | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0034063 | involved_in stress granule assembly | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0035925 | enables mRNA 3-UTR AU-rich region binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0048024 | involved_in regulation of mRNA splicing, via spliceosome | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0097165 | located_in nuclear stress granule | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:1903608 | involved_in protein localization to cytoplasmic stress granule | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:1904037 | involved_in positive regulation of epithelial cell apoptotic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:1990904 | part_of ribonucleoprotein complex | 1 | IEA | Homo_sapiens(9606) | Component |