Welcome to
RBP World!
Description
| Ensembl ID | ENSG00000115267 | Gene ID | 64135 | Accession | 18873 |
| Symbol | IFIH1 | Alias | AGS7;Hlcd;MDA5;IMD95;MDA-5;RLR-2;IDDM19;SGMRT1 | Full Name | interferon induced with helicase C domain 1 |
| Status | Confidence | Length | 51611 bases | Strand | Minus strand |
| Position | 2 : 162267074 - 162318684 | RNA binding domain | ResIII , DEAD , RIG-I_C , RIG-I_C-RD | ||
| Summary | IFIH1 encodes MDA5 which is an intracellular sensor of viral RNA that triggers the innate immune response. Sensing RNA length and secondary structure, MDA5 binds dsRNA oligonucleotides with a modified DExD/H-box helicase core and a C-terminal domain, thus leading to a proinflammatory response that includes interferons. It has been shown that Coronaviruses (CoVs) as well as various other virus families, are capable of evading the MDA5-dependent interferon response, thus impeding the activation of the innate immune response to infection. MDA5 has also been shown to play an important role in enhancing natural killer cell function in malaria infection. In addition to its protective role in antiviral responses, MDA5 has been implicated in autoimmune and autoinflammatory diseases such as type 1 diabetes, systemic lupus erythematosus, and Aicardi-Goutieres syndrome[provided by RefSeq, Jul 2020] | ||||
RNA binding domains (RBDs)
| Protein | Domain | Pfam ID | E-value | Domain number |
|---|---|---|---|---|
| ENSP00000505518 | DEAD | PF00270 | 9.8e-18 | 1 |
| ENSP00000505518 | ResIII | PF04851 | 4.5e-20 | 2 |
| ENSP00000505518 | RIG-I_C-RD | PF11648 | 2.8e-35 | 3 |
| ENSP00000505518 | RIG-I_C | PF18119 | 6.2e-27 | 4 |
| ENSP00000496816 | DEAD | PF00270 | 1.1e-17 | 1 |
| ENSP00000496816 | ResIII | PF04851 | 3.9e-20 | 2 |
| ENSP00000496816 | RIG-I_C-RD | PF11648 | 3.1e-35 | 3 |
| ENSP00000496816 | RIG-I_C | PF18119 | 1.2e-25 | 4 |
| ENSP00000497271 | DEAD | PF00270 | 1.2e-17 | 1 |
| ENSP00000497271 | ResIII | PF04851 | 4.1e-20 | 2 |
| ENSP00000497271 | RIG-I_C-RD | PF11648 | 3.3e-35 | 3 |
| ENSP00000497271 | RIG-I_C | PF18119 | 7e-27 | 4 |
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|---|---|---|---|
| 20644636 | Study of transcriptional effects in Cis at the IFIH1 locus. | Hana Zouk | 2010-07-13 | PloS one |
| 34758847 | Functional screen of inflammatory bowel disease genes reveals key epithelial functions. | Carol Jessy Ntunzwenimana | 2021-11-11 | Genome medicine |
| 33483996 | De-coding genetic risk variants in type 1 diabetes. | R Melanie Shapiro | 2021-05-01 | Immunology and cell biology |
| 35506564 | From bats to pangolins: new insights into species differences in the structure and function of the immune system. | J Patrick Haley | 2022-04-01 | Innate immunity |
| 23441136 | Admixture mapping in lupus identifies multiple functional variants within IFIH1 associated with apoptosis, inflammation, and autoantibody production. | E Julio Molineros | 2013-01-01 | PLoS genetics |
| 35646086 | RNA Helicases in Microsatellite Repeat Expansion Disorders and Neurodegeneration. | M Lydia Castelli | 2022-01-01 | Frontiers in genetics |
| 26573543 | Genetics of Lupus Nephritis: Clinical Implications. | E Melissa Munroe | 2015-09-01 | Seminars in nephrology |
| 25579795 | The RIG-I-like helicase receptor MDA5 (IFIH1) is involved in the host defense against Candida infections. | Martin Jaeger | 2015-05-01 | European journal of clinical microbiology & infectious diseases : official publication of the European Society of Clinical Microbiology |
| 26554915 | Autoimmunity in picornavirus infections. | Chandirasegaran Massilamany | 2016-02-01 | Current opinion in virology |
| 30201512 | A Balancing Act: MDA5 in Antiviral Immunity and Autoinflammation. | Gregorio Antonio AG Dias | 2019-01-01 | Trends in microbiology |
| 31898846 | Genetic and phenotypic spectrum associated with IFIH1 gain-of-function. | I Gillian Rice | 2020-04-01 | Human mutation |
| 26196376 | Newly recognized Mendelian disorders with rheumatic manifestations. | Almeida Adriana AA de | 2015-09-01 | Current opinion in rheumatology |
| 24686847 | Gain-of-function mutations in IFIH1 cause a spectrum of human disease phenotypes associated with upregulated type I interferon signaling. | I Gillian Rice | 2014-05-01 | Nature genetics |
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| IFIH1-208 | ENST00000697291 | 3712 | 177aa | ENSP00000513228 |
| IFIH1-206 | ENST00000649979 | 3581 | 1025aa | ENSP00000497271 |
| IFIH1-207 | ENST00000679938 | 2964 | 921aa | ENSP00000505518 |
| IFIH1-205 | ENST00000649554 | 2795 | No protein | - |
| IFIH1-203 | ENST00000648433 | 3234 | 986aa | ENSP00000496816 |
| IFIH1-204 | ENST00000649426 | 791 | No protein | - |
| IFIH1-202 | ENST00000464129 | 567 | No protein | - |
| IFIH1-201 | ENST00000421365 | 1617 | 221aa | ENSP00000408450 |
Phenotypes
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| 3226 | Cholesterol | 5.48668531993075E-8 | 17903299 |
| 3228 | Cholesterol | 3.92045195386449E-8 | 17903299 |
| 4644 | Cholesterol | 7.47812602937582E-7 | 17903299 |
| 60712 | Autoantibodies | 2E-14 | 21829393 |
| 60713 | Diabetes Mellitus, Type 1 | 2E-14 | 21829393 |
| 69022 | Inflammatory Bowel Diseases | 4E-10 | 26192919 |
| 70425 | Psoriasis | 1E-18 | 25903422 |
| 76066 | Diabetes Mellitus, Type 1 | 2E-11 | 17554260 |
| 88491 | Psoriasis | 1E-7 | 25903422 |
| 93828 | Lupus Erythematosus, Systemic | 4E-8 | 26502338 |
| 95550 | Psoriasis | 3E-11 | 25903422 |
| 102542 | Proteins | 7E-10 | 20694011 |
| 102605 | Psoriasis | 1E-8 | 25903422 |
| 105733 | Diabetes Mellitus, Type 1 | 7E-9 | 19430480 |
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
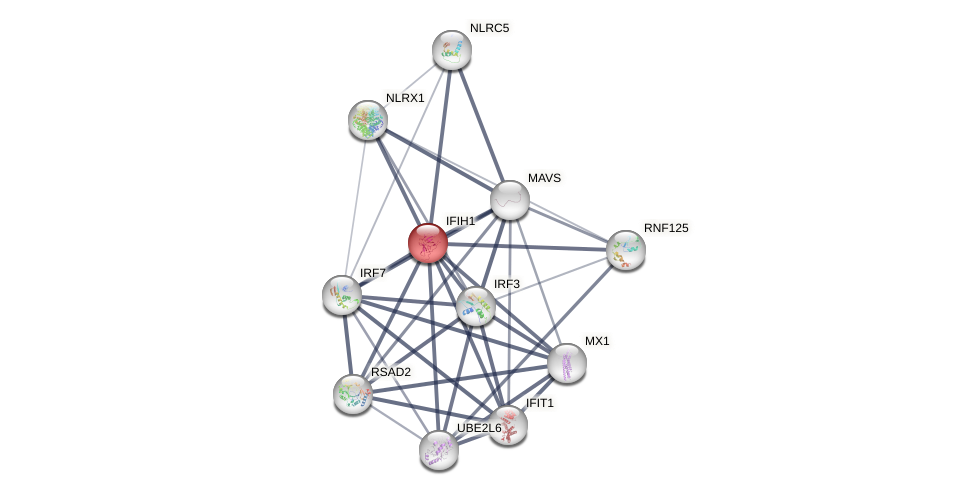
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000115267 | homo_sapiens | ENSP00000251642 | 61.0619 | 42.4779 | homo_sapiens | ENSP00000497271 | 40.3902 | 28.0976 |
| ENSG00000115267 | homo_sapiens | ENSP00000369213 | 49.2973 | 32.7568 | homo_sapiens | ENSP00000497271 | 44.4878 | 29.561 |
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 94.439 | 93.8537 | nomascus_leucogenys | ENSNLEP00000023306 | 99.1803 | 98.5656 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 99.7073 | 99.6098 | pan_paniscus | ENSPPAP00000031468 | 99.7073 | 99.6098 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 99.1219 | 98.439 | pongo_abelii | ENSPPYP00000014381 | 99.1219 | 98.439 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 99.7073 | 99.5122 | pan_troglodytes | ENSPTRP00000021498 | 99.7073 | 99.5122 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 99.7073 | 99.4146 | gorilla_gorilla | ENSGGOP00000015505 | 99.7073 | 99.4146 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 79.6098 | 68 | ornithorhynchus_anatinus | ENSOANP00000009902 | 77.6403 | 66.3178 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 83.8049 | 73.8537 | sarcophilus_harrisii | ENSSHAP00000021210 | 84.2157 | 74.2157 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 65.9512 | 59.3171 | notamacropus_eugenii | ENSMEUP00000003031 | 66.0156 | 59.375 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 66.3415 | 62.3415 | choloepus_hoffmanni | ENSCHOP00000012273 | 71.4286 | 67.1218 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 92.8781 | 86.9268 | dasypus_novemcinctus | ENSDNOP00000009395 | 92.9688 | 87.0117 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 87.0244 | 80.5854 | erinaceus_europaeus | ENSEEUP00000004719 | 87.1094 | 80.6641 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 69.3659 | 62.439 | echinops_telfairi | ENSETEP00000009142 | 70.396 | 63.3663 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 92.9756 | 89.561 | callithrix_jacchus | ENSCJAP00000069221 | 92.5243 | 89.1262 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 99.0244 | 98.1463 | cercocebus_atys | ENSCATP00000016245 | 99.0244 | 98.1463 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 99.1219 | 98.3415 | macaca_fascicularis | ENSMFAP00000034733 | 99.1219 | 98.3415 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 99.1219 | 98.2439 | macaca_nemestrina | ENSMNEP00000017489 | 99.1219 | 98.2439 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 99.0244 | 98.1463 | papio_anubis | ENSPANP00000017371 | 99.0244 | 98.1463 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 98.7317 | 98.0488 | mandrillus_leucophaeus | ENSMLEP00000000093 | 98.7317 | 98.0488 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 86.5366 | 80.0976 | tursiops_truncatus | ENSTTRP00000013071 | 86.8756 | 80.4114 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 89.7561 | 83.1219 | vulpes_vulpes | ENSVVUP00000025324 | 89.5813 | 82.9601 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 87.0244 | 80.1951 | mus_spretus | MGP_SPRETEiJ_P0054047 | 87.0244 | 80.1951 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 91.7073 | 86.1463 | equus_asinus | ENSEASP00005058075 | 91.6179 | 86.0624 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 90.6341 | 83.7073 | capra_hircus | ENSCHIP00000021583 | 90.8113 | 83.871 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 90.7317 | 84.7805 | loxodonta_africana | ENSLAFP00000008373 | 92.4453 | 86.3817 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 91.5122 | 84.8781 | panthera_leo | ENSPLOP00000019803 | 91.423 | 84.7953 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 89.8537 | 84.2927 | ailuropoda_melanoleuca | ENSAMEP00000006040 | 90.3827 | 84.789 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 90.2439 | 83.4146 | bos_indicus_hybrid | ENSBIXP00005040063 | 90.5975 | 83.7414 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 90.1463 | 84.7805 | oryctolagus_cuniculus | ENSOCUP00000027386 | 88.5057 | 83.2375 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 91.7073 | 86.1463 | equus_caballus | ENSECAP00000006532 | 91.6179 | 86.0624 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 88.7805 | 81.7561 | canis_lupus_familiaris | ENSCAFP00845042274 | 87.5842 | 80.6545 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 91.4146 | 84.9756 | panthera_pardus | ENSPPRP00000000647 | 91.3255 | 84.8928 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 92.1951 | 86.1463 | marmota_marmota_marmota | ENSMMMP00000026198 | 92.3754 | 86.3148 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 90.9268 | 85.6585 | balaenoptera_musculus | ENSBMSP00010016832 | 91.4622 | 86.1629 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 89.561 | 82.9268 | cavia_porcellus | ENSCPOP00000006451 | 89.8239 | 83.1703 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 91.2195 | 84.8781 | felis_catus | ENSFCAP00000019326 | 91.1306 | 84.7953 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 90.1463 | 84.4878 | ursus_americanus | ENSUAMP00000020969 | 90.8555 | 85.1524 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 81.9512 | 72.7805 | monodelphis_domestica | ENSMODP00000044985 | 82.2723 | 73.0656 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 90.1463 | 83.1219 | bison_bison_bison | ENSBBBP00000006213 | 90.4995 | 83.4476 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 91.6098 | 86.0488 | monodon_monoceros | ENSMMNP00015009178 | 91.7889 | 86.217 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 91.6098 | 84.6829 | sus_scrofa | ENSSSCP00000044491 | 91.7889 | 84.8485 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 92.2927 | 86.9268 | microcebus_murinus | ENSMICP00000004929 | 91.7556 | 86.421 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 89.1707 | 82.6341 | octodon_degus | ENSODEP00000018433 | 89.6958 | 83.1207 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 85.3659 | 79.0244 | jaculus_jaculus | ENSJJAP00000016972 | 89.5599 | 82.9069 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 90.6341 | 83.4146 | ovis_aries_rambouillet | ENSOARP00020013826 | 90.8113 | 83.5777 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 88.8781 | 83.0244 | ursus_maritimus | ENSUMAP00000016375 | 89.6654 | 83.7598 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 76.3902 | 71.4146 | ochotona_princeps | ENSOPRP00000014414 | 77.6786 | 72.619 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 83.7073 | 78.5366 | delphinapterus_leucas | ENSDLEP00000015511 | 91.9614 | 86.2808 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 91.4146 | 85.9512 | physeter_catodon | ENSPCTP00005019109 | 91.5934 | 86.1193 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 86.9268 | 80 | mus_caroli | MGP_CAROLIEiJ_P0051982 | 86.9268 | 80 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 87.0244 | 79.8049 | mus_musculus | ENSMUSP00000028259 | 87.0244 | 79.8049 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 90.7317 | 83.6098 | dipodomys_ordii | ENSDORP00000019151 | 91.8065 | 84.6002 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 89.7561 | 83.8049 | mustela_putorius_furo | ENSMPUP00000000558 | 89.5813 | 83.6417 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 95.8049 | 92.9756 | aotus_nancymaae | ENSANAP00000020745 | 95.8049 | 92.9756 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 95.4146 | 92.5854 | saimiri_boliviensis_boliviensis | ENSSBOP00000019200 | 95.4146 | 92.5854 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 73.4634 | 67.6098 | vicugna_pacos | ENSVPAP00000010078 | 87.052 | 80.1156 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 99.0244 | 98.2439 | chlorocebus_sabaeus | ENSCSAP00000011653 | 99.0244 | 98.2439 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 81.7561 | 76.2927 | tupaia_belangeri | ENSTBEP00000004849 | 82.3183 | 76.8173 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 91.8049 | 85.6585 | phocoena_sinus | ENSPSNP00000003110 | 91.9844 | 85.826 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 91.5122 | 85.2683 | camelus_dromedarius | ENSCDRP00005005506 | 91.7808 | 85.5186 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 93.9512 | 90.3415 | carlito_syrichta | ENSTSYP00000005394 | 93.9512 | 90.3415 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 91.2195 | 84.7805 | panthera_tigris_altaica | ENSPTIP00000023434 | 90.6887 | 84.2871 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 87.2195 | 79.9024 | rattus_norvegicus | ENSRNOP00000009621 | 87.0497 | 79.7468 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 94.439 | 89.561 | prolemur_simus | ENSPSMP00000020951 | 94.439 | 89.561 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 87.6098 | 80.7805 | microtus_ochrogaster | ENSMOCP00000011326 | 88.2989 | 81.4159 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 61.2683 | 55.5122 | sorex_araneus | ENSSARP00000002336 | 61.5083 | 55.7297 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 87.9024 | 81.0732 | peromyscus_maniculatus_bairdii | ENSPEMP00000014929 | 88.1605 | 81.3112 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 89.0732 | 82.3415 | bos_mutus | ENSBMUP00000023828 | 89.5976 | 82.8263 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 87.0244 | 80.0976 | mus_spicilegus | ENSMSIP00000024181 | 87.0244 | 80.0976 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 87.5122 | 80.9756 | cricetulus_griseus_chok1gshd | ENSCGRP00001027862 | 87.7691 | 81.2133 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 87.3171 | 81.2683 | pteropus_vampyrus | ENSPVAP00000002497 | 88.1773 | 82.069 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 90.3415 | 83.4146 | myotis_lucifugus | ENSMLUP00000008664 | 90.6067 | 83.6595 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 84.6829 | 75.4146 | vombatus_ursinus | ENSVURP00010028127 | 84.518 | 75.2678 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 90.8293 | 84.4878 | rhinolophus_ferrumequinum | ENSRFEP00010007227 | 90.2132 | 83.9147 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 89.7561 | 83.7073 | neovison_vison | ENSNVIP00000008306 | 89.9316 | 83.871 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 90.3415 | 83.4146 | bos_taurus | ENSBTAP00000010703 | 90.6954 | 83.7414 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 89.4634 | 82.8293 | heterocephalus_glaber_female | ENSHGLP00000012269 | 89.2892 | 82.668 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 95.8049 | 94.7317 | rhinopithecus_bieti | ENSRBIP00000010720 | 96.8442 | 95.7594 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 89.9512 | 83.3171 | canis_lupus_dingo | ENSCAFP00020005650 | 89.6016 | 82.9932 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 91.4146 | 86.1463 | sciurus_vulgaris | ENSSVLP00005017680 | 91.4146 | 86.1463 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 83.9024 | 74.7317 | phascolarctos_cinereus | ENSPCIP00000041521 | 83.6576 | 74.5136 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 77.9512 | 71.6098 | procavia_capensis | ENSPCAP00000007183 | 85.6377 | 78.671 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 89.6585 | 83.2195 | nannospalax_galili | ENSNGAP00000010303 | 90.0098 | 83.5455 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 90.0488 | 83.3171 | bos_grunniens | ENSBGRP00000027560 | 90.4016 | 83.6435 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 89.8537 | 84.0976 | chinchilla_lanigera | ENSCLAP00000012317 | 89.7661 | 84.0156 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 89.8537 | 83.5122 | cervus_hanglu_yarkandensis | ENSCHYP00000016626 | 90.2941 | 83.9216 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 91.3171 | 84.1951 | catagonus_wagneri | ENSCWAP00000002390 | 91.4956 | 84.3597 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 88.6829 | 82.8293 | ictidomys_tridecemlineatus | ENSSTOP00000004468 | 93.3265 | 87.1663 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 91.5122 | 85.3659 | otolemur_garnettii | ENSOGAP00000011298 | 91.6016 | 85.4492 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 95.2195 | 92.3902 | cebus_imitator | ENSCCAP00000018819 | 95.2195 | 92.3902 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 92.1951 | 85.9512 | urocitellus_parryii | ENSUPAP00010014102 | 87.9888 | 82.0298 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 90.3415 | 83.0244 | moschus_moschiferus | ENSMMSP00000032042 | 90.6067 | 83.2681 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 98.9268 | 98.1463 | rhinopithecus_roxellana | ENSRROP00000013435 | 98.9268 | 98.1463 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 86.7317 | 79.4146 | mus_pahari | MGP_PahariEiJ_P0062975 | 86.7317 | 79.4146 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 93.561 | 87.8049 | propithecus_coquereli | ENSPCOP00000019450 | 93.561 | 87.8049 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 39.9024 | 21.9512 | caenorhabditis_elegans | D2005.5.1 | 36.5505 | 20.1072 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 36.7805 | 20.7805 | caenorhabditis_elegans | F15B10.2a.1 | 36.3549 | 20.54 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 15.3171 | 7.90244 | ciona_intestinalis | ENSCINP00000023858 | 38.2927 | 19.7561 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 28.9756 | 21.6585 | petromyzon_marinus | ENSPMAP00000008095 | 64.4252 | 48.1562 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 34.439 | 25.8537 | eptatretus_burgeri | ENSEBUP00000019471 | 59.6284 | 44.7635 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 66.7317 | 51.5122 | callorhinchus_milii | ENSCMIP00000016812 | 68.5371 | 52.9058 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 56.0976 | 47.0244 | latimeria_chalumnae | ENSLACP00000012995 | 78.6594 | 65.9371 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 68.0976 | 54.1463 | lepisosteus_oculatus | ENSLOCP00000010298 | 68.7685 | 54.6798 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 59.7073 | 43.6098 | clupea_harengus | ENSCHAP00000014285 | 51.2563 | 37.4372 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 62.7317 | 46.9268 | clupea_harengus | ENSCHAP00000062558 | 65.2792 | 48.8325 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 63.7073 | 46.9268 | danio_rerio | ENSDARP00000013943 | 65.4965 | 48.2447 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 61.6585 | 46.7317 | carassius_auratus | ENSCARP00000003902 | 63.8384 | 48.3838 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 32 | 21.8537 | carassius_auratus | ENSCARP00000080572 | 54.2149 | 37.0248 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 62.5366 | 46.9268 | astyanax_mexicanus | ENSAMXP00000010301 | 62.7202 | 47.0646 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 62.6341 | 47.9024 | ictalurus_punctatus | ENSIPUP00000019810 | 63.8806 | 48.8557 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 63.0244 | 47.3171 | esox_lucius | ENSELUP00000020430 | 65.0554 | 48.8419 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 63.3171 | 46.9268 | electrophorus_electricus | ENSEEEP00000050306 | 64.2574 | 47.6238 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 62.9268 | 47.122 | oncorhynchus_kisutch | ENSOKIP00005070318 | 64.5 | 48.3 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 63.122 | 47.2195 | oncorhynchus_mykiss | ENSOMYP00000069679 | 64.5709 | 48.3034 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 62.8293 | 47.4146 | salmo_salar | ENSSSAP00000113266 | 63.0754 | 47.6004 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 63.2195 | 47.2195 | salmo_trutta | ENSSTUP00000034572 | 64.3496 | 48.0636 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 60.6829 | 43.9024 | gadus_morhua | ENSGMOP00000031164 | 63.0192 | 45.5927 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 60.7805 | 45.2683 | fundulus_heteroclitus | ENSFHEP00000034810 | 64.0288 | 47.6876 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 60.878 | 44 | poecilia_reticulata | ENSPREP00000011433 | 64.3963 | 46.5428 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 60.878 | 44.878 | xiphophorus_maculatus | ENSXMAP00000011583 | 64.3299 | 47.4227 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 61.7561 | 44.6829 | oryzias_latipes | ENSORLP00000021147 | 64.9897 | 47.0226 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 60.5854 | 44.6829 | cyclopterus_lumpus | ENSCLMP00005006804 | 63.2383 | 46.6395 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 62.6341 | 46.439 | oreochromis_niloticus | ENSONIP00000059635 | 61.9691 | 45.9459 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 62.6341 | 46.3415 | haplochromis_burtoni | ENSHBUP00000003561 | 64.7177 | 47.8831 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 62.5366 | 46.5366 | astatotilapia_calliptera | ENSACLP00000019012 | 64.6169 | 48.0847 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 61.7561 | 46.1463 | sparus_aurata | ENSSAUP00010055285 | 63.8105 | 47.6814 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 61.9512 | 46.9268 | lates_calcarifer | ENSLCAP00010056143 | 64.9284 | 49.182 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 68.0976 | 54.1463 | xenopus_tropicalis | ENSXETP00000085398 | 69.8699 | 55.5556 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 75.8049 | 64.6829 | chrysemys_picta_bellii | ENSCPBP00000038200 | 76.9307 | 65.6436 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 73.561 | 62.2439 | sphenodon_punctatus | ENSSPUP00000022629 | 75.2495 | 63.6727 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 70.7317 | 59.5122 | crocodylus_porosus | ENSCPRP00005026760 | 73.3806 | 61.7409 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 48.5854 | 39.8049 | laticauda_laticaudata | ENSLLTP00000020918 | 68.2192 | 55.8904 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 65.3659 | 53.6585 | notechis_scutatus | ENSNSUP00000028963 | 67.0671 | 55.0551 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 64.1951 | 53.0732 | notechis_scutatus | ENSNSUP00000028841 | 68.9727 | 57.0231 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 66.7317 | 54.8293 | pseudonaja_textilis | ENSPTXP00000012579 | 67.2566 | 55.2606 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 73.1707 | 61.561 | anas_platyrhynchos_platyrhynchos | ENSAPLP00000010681 | 74.7757 | 62.9113 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 72.0976 | 60.1951 | gallus_gallus | ENSGALP00010029975 | 67.0599 | 55.9891 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 57.8537 | 51.0244 | meleagris_gallopavo | ENSMGAP00000029088 | 79.8116 | 70.3903 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 70.2439 | 59.8049 | serinus_canaria | ENSSCAP00000020623 | 72.0721 | 61.3614 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 73.0732 | 61.0732 | parus_major | ENSPMJP00000026930 | 74.3793 | 62.1648 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 69.4634 | 58.5366 | anolis_carolinensis | ENSACAP00000013841 | 70.7753 | 59.6421 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 62.3415 | 46.439 | neolamprologus_brichardi | ENSNBRP00000019203 | 65.5385 | 48.8205 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 48.1951 | 40.5854 | naja_naja | ENSNNAP00000007043 | 75.3049 | 63.4146 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 69.2683 | 57.9512 | podarcis_muralis | ENSPMRP00000017604 | 71.1423 | 59.519 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 67.8049 | 56.4878 | geospiza_fortis | ENSGFOP00000015121 | 71.1361 | 59.2631 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 72.2927 | 60.3902 | taeniopygia_guttata | ENSTGUP00000007146 | 73.6581 | 61.5308 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 65.8537 | 50.5366 | erpetoichthys_calabaricus | ENSECRP00000008922 | 67.5 | 51.8 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 61.9512 | 45.8537 | kryptolebias_marmoratus | ENSKMAP00000019253 | 65.2621 | 48.3042 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 68.8781 | 56.9756 | salvator_merianae | ENSSMRP00000030336 | 70.5295 | 58.3417 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 61.561 | 44.878 | oryzias_javanicus | ENSOJAP00000027480 | 63.9959 | 46.6531 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 60.6829 | 45.9512 | amphilophus_citrinellus | ENSACIP00000027361 | 63.0192 | 47.7204 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 63.7073 | 48.1951 | dicentrarchus_labrax | ENSDLAP00005045388 | 66.0263 | 49.9494 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 60.878 | 45.6585 | amphiprion_percula | ENSAPEP00000026557 | 64.1316 | 48.0987 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 61.7561 | 44.9756 | oryzias_sinensis | ENSOSIP00000039904 | 64.9897 | 47.3306 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 75.3171 | 64.0976 | pelodiscus_sinensis | ENSPSIP00000008310 | 75.835 | 64.5383 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 68.1951 | 57.9512 | gopherus_evgoodei | ENSGEVP00005028810 | 73.7342 | 62.6582 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 75.6098 | 64.2927 | chelonoidis_abingdonii | ENSCABP00000008310 | 76.1297 | 64.7348 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 62.7317 | 47.4146 | seriola_dumerili | ENSSDUP00000026204 | 65.3455 | 49.3902 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 60.9756 | 46.3415 | cottoperca_gobio | ENSCGOP00000048247 | 63.9713 | 48.6182 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 70.2439 | 59.0244 | struthio_camelus_australis | ENSSCUP00000013572 | 72.1443 | 60.6212 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 73.561 | 61.9512 | anser_brachyrhynchus | ENSABRP00000026053 | 75.0996 | 63.247 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 61.0732 | 46.3415 | gasterosteus_aculeatus | ENSGACP00000007327 | 62.1032 | 47.123 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 59.122 | 43.122 | cynoglossus_semilaevis | ENSCSEP00000016067 | 63.7224 | 46.4774 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 60.3902 | 44.3902 | poecilia_formosa | ENSPFOP00000017315 | 63.8144 | 46.9072 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 61.2683 | 44.7805 | myripristis_murdjan | ENSMMDP00005054392 | 65.0104 | 47.5155 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 62.439 | 46.2439 | sander_lucioperca | ENSSLUP00000040968 | 65.7084 | 48.6653 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 72.2927 | 60.7805 | strigops_habroptila | ENSSHBP00005024342 | 73.8048 | 62.0518 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 60.9756 | 45.7561 | amphiprion_ocellaris | ENSAOCP00000010559 | 64.2343 | 48.2014 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 76.1951 | 65.0732 | terrapene_carolina_triunguis | ENSTMTP00000031870 | 77.3267 | 66.0396 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 63.0244 | 47.122 | hucho_hucho | ENSHHUP00000003998 | 64.0238 | 47.8692 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 61.7561 | 44.3902 | betta_splendens | ENSBSLP00000047477 | 64.6578 | 46.476 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 63.122 | 47.8049 | pygocentrus_nattereri | ENSPNAP00000034485 | 63.556 | 48.1336 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 62.9268 | 45.561 | anabas_testudineus | ENSATEP00000026498 | 65.6155 | 47.5076 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 6.34146 | 3.90244 | ciona_savignyi | ENSCSAVP00000018923 | 33.6788 | 20.7254 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 3.90244 | 2.14634 | saccharomyces_cerevisiae | YHR218W-A | 38.0952 | 20.9524 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 25.7561 | 12.4878 | saccharomyces_cerevisiae | YOR396W | 14.6993 | 7.12695 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 9.56098 | 4.87805 | saccharomyces_cerevisiae | YHL050C | 14.0603 | 7.1736 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 14.7317 | 7.80488 | saccharomyces_cerevisiae | YPR204W | 14.6318 | 7.75194 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 18.5366 | 9.65854 | saccharomyces_cerevisiae | YLL066C | 15.7676 | 8.21577 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 13.3659 | 6.34146 | saccharomyces_cerevisiae | YHR218W | 22.7197 | 10.7794 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 25.7561 | 12.5854 | saccharomyces_cerevisiae | YJL225C | 15.0171 | 7.33788 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 28.3902 | 13.8537 | saccharomyces_cerevisiae | YNL339C | 15.6536 | 7.63852 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 25.7561 | 12.4878 | saccharomyces_cerevisiae | YLR467W | 14.6993 | 7.12695 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 6.14634 | 2.92683 | saccharomyces_cerevisiae | YBL113C | 7.95455 | 3.78788 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 25.7561 | 12.4878 | saccharomyces_cerevisiae | YDR545W | 14.6993 | 7.12695 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 25.7561 | 12.5854 | saccharomyces_cerevisiae | YIL177C | 15.0171 | 7.33788 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 7.90244 | 3.60976 | saccharomyces_cerevisiae | YPR202W | 34.0336 | 15.5462 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 8.97561 | 5.07317 | saccharomyces_cerevisiae | YFL066C | 23.4694 | 13.2653 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 4.29268 | 2.14634 | saccharomyces_cerevisiae | YEL075C | 36.0656 | 18.0328 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 17.561 | 8.87805 | saccharomyces_cerevisiae | YEL077C | 14.0955 | 7.12608 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 21.561 | 10.5366 | saccharomyces_cerevisiae | YER190W | 13.1469 | 6.42475 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 6.2439 | 3.02439 | saccharomyces_cerevisiae | YLR462W | 31.6832 | 15.3465 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 3.90244 | 2.14634 | saccharomyces_cerevisiae | YBL112C | 38.0952 | 20.9524 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 28.3902 | 13.8537 | saccharomyces_cerevisiae | YPL283C | 15.6536 | 7.63852 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 7.41463 | 3.41463 | saccharomyces_cerevisiae | YHL049C | 28.0443 | 12.9151 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 24.1951 | 12.1951 | saccharomyces_cerevisiae | YML133C | 18.0495 | 9.09753 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 6.2439 | 3.12195 | saccharomyces_cerevisiae | YHR219W | 10.2564 | 5.12821 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 28.3902 | 13.8537 | saccharomyces_cerevisiae | YGR296W | 15.6536 | 7.63852 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 18.3415 | 9.46341 | saccharomyces_cerevisiae | YLL067C | 15.6017 | 8.04979 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 18.1463 | 9.07317 | saccharomyces_cerevisiae | YLR466W | 13.4588 | 6.72938 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 4.29268 | 2.04878 | saccharomyces_cerevisiae | YER189W | 36.0656 | 17.2131 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 15.4146 | 7.5122 | saccharomyces_cerevisiae | YBL111C | 23.6882 | 11.5442 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 5.36585 | 2.43902 | saccharomyces_cerevisiae | YFL064C | 31.6092 | 14.3678 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 62.5366 | 47.2195 | seriola_lalandi_dorsalis | ENSSLDP00000020910 | 65.1423 | 49.187 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 61.561 | 44.2927 | labrus_bergylta | ENSLBEP00000006965 | 63.7374 | 45.8586 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 62.7317 | 46.3415 | pundamilia_nyererei | ENSPNYP00000003897 | 65.412 | 48.3215 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 62.8293 | 47.122 | mastacembelus_armatus | ENSMAMP00000005039 | 64.9849 | 48.7386 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 57.9512 | 44.1951 | stegastes_partitus | ENSSPAP00000001115 | 64.1469 | 48.9201 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 63.122 | 47.0244 | oncorhynchus_tshawytscha | ENSOTSP00005089227 | 64.5709 | 48.1038 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 61.6585 | 46.6341 | acanthochromis_polyacanthus | ENSAPOP00000020900 | 64.8871 | 49.076 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 73.7561 | 62.1463 | aquila_chrysaetos_chrysaetos | ENSACCP00020016119 | 72.1374 | 60.7824 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 60.3902 | 44.9756 | nothobranchius_furzeri | ENSNFUP00015044322 | 61.592 | 45.8706 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 61.6585 | 46.6341 | cyprinodon_variegatus | ENSCVAP00000001133 | 65.0875 | 49.2276 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 59.2195 | 42.1463 | denticeps_clupeoides | ENSDCDP00000046311 | 59.6267 | 42.4361 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 62.2439 | 46.439 | denticeps_clupeoides | ENSDCDP00000038389 | 64.2497 | 47.9356 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 62.9268 | 46.8293 | larimichthys_crocea | ENSLCRP00005063006 | 65.0858 | 48.4359 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 59.4146 | 43.4146 | takifugu_rubripes | ENSTRUP00000003254 | 62.8483 | 45.9236 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 72.0976 | 60.2927 | ficedula_albicollis | ENSFALP00000000746 | 72.8079 | 60.8867 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 59.3171 | 42.8293 | hippocampus_comes | ENSHCOP00000025724 | 63.0705 | 45.5394 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 61.8537 | 47.122 | cyprinus_carpio_carpio | ENSCCRP00000025616 | 63.7827 | 48.5915 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 61.9512 | 46.7317 | cyprinus_carpio_carpio | ENSCCRP00000117543 | 64.4016 | 48.5801 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 71.4146 | 60.2927 | coturnix_japonica | ENSCJPP00005019356 | 67.2794 | 56.8015 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 60.2927 | 44.3902 | poecilia_latipinna | ENSPLAP00000019102 | 63.7113 | 46.9072 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 62.1463 | 47.0244 | sinocyclocheilus_grahami | ENSSGRP00000098675 | 64.4737 | 48.7854 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 59.7073 | 44.3902 | sinocyclocheilus_grahami | ENSSGRP00000058182 | 62.5767 | 46.5235 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 62.5366 | 46.439 | maylandia_zebra | ENSMZEP00005009445 | 64.6169 | 47.9839 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 59.9024 | 42.9268 | tetraodon_nigroviridis | ENSTNIP00000019599 | 61.2774 | 43.9122 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 61.8537 | 47.8049 | paramormyrops_kingsleyae | ENSPKIP00000013751 | 62.8345 | 48.5629 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 64.1951 | 49.4634 | scleropages_formosus | ENSSFOP00015035662 | 63.2692 | 48.75 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 66.5366 | 51.5122 | leptobrachium_leishanense | ENSLLEP00000047037 | 66.2136 | 51.2621 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 61.561 | 45.7561 | scophthalmus_maximus | ENSSMAP00000020234 | 62.9741 | 46.8064 |
| ENSG00000115267 | homo_sapiens | ENSP00000497271 | 62.3415 | 45.2683 | oryzias_melastigma | ENSOMEP00000022778 | 65.8084 | 47.7858 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0002753 | involved_in cytosolic pattern recognition receptor signaling pathway | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0003677 | enables DNA binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0003723 | enables RNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0003724 | enables RNA helicase activity | 1 | IMP | Homo_sapiens(9606) | Function |
| GO:0003725 | enables double-stranded RNA binding | 3 | IBA,IDA,TAS | Homo_sapiens(9606) | Function |
| GO:0003727 | enables single-stranded RNA binding | 2 | IBA,IDA | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005524 | enables ATP binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0005634 | located_in nucleus | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0005737 | is_active_in cytoplasm | 3 | IBA,IDA | Homo_sapiens(9606) | Component |
| GO:0005739 | located_in mitochondrion | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005829 | located_in cytosol | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0008270 | enables zinc ion binding | 2 | IBA,IDA | Homo_sapiens(9606) | Function |
| GO:0009597 | involved_in detection of virus | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0009615 | involved_in response to virus | 2 | IDA,TAS | Homo_sapiens(9606) | Process |
| GO:0016887 | enables ATP hydrolysis activity | 1 | IMP | Homo_sapiens(9606) | Function |
| GO:0016925 | involved_in protein sumoylation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0032559 | enables adenyl ribonucleotide binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0032727 | involved_in positive regulation of interferon-alpha production | 2 | IMP,TAS | Homo_sapiens(9606) | Process |
| GO:0032728 | involved_in positive regulation of interferon-beta production | 3 | IDA,IMP,TAS | Homo_sapiens(9606) | Process |
| GO:0032755 | involved_in positive regulation of interleukin-6 production | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0032760 | involved_in positive regulation of tumor necrosis factor production | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0034344 | involved_in regulation of type III interferon production | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0035639 | enables purine ribonucleoside triphosphate binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0038187 | enables pattern recognition receptor activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0039530 | involved_in MDA-5 signaling pathway | 3 | IBA,IDA,IMP | Homo_sapiens(9606) | Process |
| GO:0042802 | enables identical protein binding | 2 | IDA,IPI | Homo_sapiens(9606) | Function |
| GO:0043021 | enables ribonucleoprotein complex binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0045071 | involved_in negative regulation of viral genome replication | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0045087 | involved_in innate immune response | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0051259 | involved_in protein complex oligomerization | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0051607 | involved_in defense response to virus | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0060337 | involved_in type I interferon-mediated signaling pathway | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0060760 | involved_in positive regulation of response to cytokine stimulus | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0071360 | involved_in cellular response to exogenous dsRNA | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0098586 | involved_in cellular response to virus | 1 | IEP | Homo_sapiens(9606) | Process |
| GO:0140374 | involved_in antiviral innate immune response | 1 | IBA | Homo_sapiens(9606) | Process |