Welcome to
RBP World!
Functions and Diseases
| RBP Type | Canonical_RBPs |
| Diseases | CancerSARS-COV-OC43FlavivirusesHIVRVSARS-COV-2SINVDengueZika |
| Drug | N.A. |
| Main interacting RNAs | mRNA |
| Moonlighting functions | Transmemebrane |
| Localizations | Stress granucle |
| BulkPerturb-seq | DataSet_01_136 DataSet_01_180 |
Description
| Ensembl ID | ENSG00000114416 | Gene ID | 8087 | Accession | 4023 |
| Symbol | FXR1 | Alias | FXR1P;CMYP9A;CMYP9B;MYOPMIL;MYORIBF | Full Name | FMR1 autosomal homolog 1 |
| Status | Confidence | Length | 114613 bases | Strand | Plus strand |
| Position | 3 : 180868141 - 180982753 | RNA binding domain | Agenet , FXR_C1 , KH_1 , KH_9 , FXMRP1_C_core | ||
| Summary | The protein encoded by this gene is an RNA binding protein that interacts with the functionally-similar proteins FMR1 and FXR2. These proteins shuttle between the nucleus and cytoplasm and associate with polyribosomes, predominantly with the 60S ribosomal subunit. Three transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jul 2008] | ||||
RNA binding domains (RBDs)
| Protein | Domain | Pfam ID | E-value | Domain number |
|---|---|---|---|---|
| ENSP00000388828 | Agenet | PF05641 | 2.3e-14 | 1 |
| ENSP00000388828 | Agenet | PF05641 | 2.3e-14 | 2 |
| ENSP00000388828 | KH_9 | PF17904 | 3.5e-36 | 3 |
| ENSP00000388828 | KH_1 | PF00013 | 3.8e-16 | 4 |
| ENSP00000388828 | KH_1 | PF00013 | 3.8e-16 | 5 |
| ENSP00000388828 | FXMRP1_C_core | PF12235 | 2e-33 | 6 |
| ENSP00000388828 | FXMRP1_C_core | PF12235 | 2e-33 | 7 |
| ENSP00000388828 | FXR_C1 | PF16096 | 1.2e-38 | 8 |
| ENSP00000513936 | Agenet | PF05641 | 2.7e-14 | 1 |
| ENSP00000513936 | Agenet | PF05641 | 2.7e-14 | 2 |
| ENSP00000513936 | KH_9 | PF17904 | 3.9e-36 | 3 |
| ENSP00000513936 | KH_1 | PF00013 | 4.4e-16 | 4 |
| ENSP00000513936 | KH_1 | PF00013 | 4.4e-16 | 5 |
| ENSP00000513936 | FXMRP1_C_core | PF12235 | 9e-54 | 6 |
| ENSP00000513936 | FXR_C1 | PF16096 | 1.3e-38 | 7 |
| ENSP00000350170 | Agenet | PF05641 | 3.6e-14 | 1 |
| ENSP00000350170 | Agenet | PF05641 | 3.6e-14 | 2 |
| ENSP00000350170 | KH_9 | PF17904 | 3.6e-36 | 3 |
| ENSP00000350170 | KH_1 | PF00013 | 5e-16 | 4 |
| ENSP00000350170 | FXMRP1_C_core | PF12235 | 3.8e-33 | 5 |
| ENSP00000350170 | FXMRP1_C_core | PF12235 | 3.8e-33 | 6 |
| ENSP00000350170 | FXR_C1 | PF16096 | 1.3e-38 | 7 |
| ENSP00000513937 | Agenet | PF05641 | 4.1e-14 | 1 |
| ENSP00000513937 | KH_9 | PF17904 | 3.9e-36 | 2 |
| ENSP00000513937 | KH_1 | PF00013 | 5.7e-16 | 3 |
| ENSP00000513937 | FXMRP1_C_core | PF12235 | 1.1e-53 | 4 |
| ENSP00000513937 | FXR_C1 | PF16096 | 1.4e-38 | 5 |
| ENSP00000417513 | Agenet | PF05641 | 2.4e-12 | 1 |
| ENSP00000417513 | KH_9 | PF17904 | 1.7e-06 | 2 |
| ENSP00000418097 | Agenet | PF05641 | 3.3e-10 | 1 |
| ENSP00000418097 | KH_9 | PF17904 | 3.5e-36 | 2 |
| ENSP00000418097 | KH_1 | PF00013 | 4.9e-16 | 3 |
| ENSP00000418097 | KH_1 | PF00013 | 4.9e-16 | 4 |
| ENSP00000418097 | FXMRP1_C_core | PF12235 | 4.2e-33 | 5 |
| ENSP00000418097 | FXMRP1_C_core | PF12235 | 4.2e-33 | 6 |
| ENSP00000418097 | FXR_C1 | PF16096 | 1.3e-38 | 7 |
| ENSP00000420643 | Agenet | PF05641 | 1.8e-05 | 1 |
| ENSP00000420643 | KH_9 | PF17904 | 3.1e-36 | 2 |
| ENSP00000420643 | KH_1 | PF00013 | 3.1e-16 | 3 |
| ENSP00000420643 | KH_1 | PF00013 | 3.1e-16 | 4 |
| ENSP00000420643 | FXMRP1_C_core | PF12235 | 1.5e-33 | 5 |
| ENSP00000420643 | FXMRP1_C_core | PF12235 | 1.5e-33 | 6 |
| ENSP00000420643 | FXR_C1 | PF16096 | 1.1e-38 | 7 |
| ENSP00000419933 | KH_9 | PF17904 | 2.5e-37 | 1 |
| ENSP00000420515 | KH_9 | PF17904 | 2.8e-36 | 1 |
| ENSP00000420515 | KH_1 | PF00013 | 2.6e-16 | 2 |
| ENSP00000420515 | KH_1 | PF00013 | 2.6e-16 | 3 |
| ENSP00000420515 | FXMRP1_C_core | PF12235 | 1.2e-33 | 4 |
| ENSP00000420515 | FXMRP1_C_core | PF12235 | 1.2e-33 | 5 |
| ENSP00000420515 | FXR_C1 | PF16096 | 9.5e-39 | 6 |
| ENSP00000307633 | KH_9 | PF17904 | 2.9e-36 | 1 |
| ENSP00000307633 | KH_1 | PF00013 | 3.6e-16 | 2 |
| ENSP00000307633 | KH_1 | PF00013 | 3.6e-16 | 3 |
| ENSP00000307633 | FXMRP1_C_core | PF12235 | 3e-32 | 4 |
| ENSP00000307633 | FXMRP1_C_core | PF12235 | 3e-32 | 5 |
| ENSP00000307633 | FXR_C1 | PF16096 | 1.1e-38 | 6 |
| ENSP00000419793 | KH_9 | PF17904 | 2.6e-35 | 1 |
| ENSP00000418724 | KH_9 | PF17904 | 2.7e-18 | 1 |
| ENSP00000417125 | KH_9 | PF17904 | 1.9e-13 | 1 |
| ENSP00000418027 | FXMRP1_C_core | PF12235 | 1.5e-13 | 1 |
| ENSP00000418027 | FXR_C1 | PF16096 | 7e-39 | 2 |
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|---|---|---|---|
| 35908416 | Intestinal epithelial β Klotho is a critical protective factor in alcohol-induced intestinal barrier dysfunction and liver injury. | Zhengping Hou | 2022-08-01 | EBioMedicine |
| 36245599 | Skeletal muscle and related protein expression as prognostic factors in thymic squamous cell carcinoma. | Keita Nakanishi | 2022-09-01 | Journal of thoracic disease |
| 32313943 | Plasticity of nuclear and cytoplasmic stress responses of RNA-binding proteins. | Michael Backlund | 2020-05-21 | Nucleic acids research |
| 36498797 | Screening and Identification of Key Biomarkers in Metastatic Uveal Melanoma: Evidence from a Bioinformatic Analysis. | Tan Wang | 2022-12-05 | Journal of clinical medicine |
| 32193154 | Upregulation of OIP5-AS1 Predicts Poor Prognosis and Contributes to Thyroid Cancer Cell Proliferation and Migration. | Qiuli Li | 2020-06-05 | Molecular therapy. Nucleic acids |
| 35886008 | Identification of Antitumor miR-30e-5p Controlled Genes; Diagnostic and Prognostic Biomarkers for Head and Neck Squamous Cell Carcinoma. | Chikashi Minemura | 2022-07-09 | Genes |
| 21072162 | Structural studies of the tandem Tudor domains of fragile X mental retardation related proteins FXR1 and FXR2. | A Melanie Adams-Cioaba | 2010-11-02 | PloS one |
| 36749074 | Fragile X-Related Protein FXR1 Controls Human Adenovirus Capsid mRNA Metabolism. | Zamaneh Hajikhezri | 2023-02-28 | Journal of virology |
| 35666183 | Integrative analysis and prediction of human R-loop binding proteins. | Arun Kumar | 2022-07-29 | G3 (Bethesda, Md.) |
| 36306353 | Ribosome changes reprogram translation for chemosurvival in G0 leukemic cells. | Chandreyee Datta | 2022-10-28 | Science advances |
| 7781595 | FXR1, an autosomal homolog of the fragile X mental retardation gene. | C M Siomi | 1995-06-01 | The EMBO journal |
| 37180732 | Study liquid-liquid phase separation with optical microscopy: A methodology review. | Xiufeng Zhang | 2023-06-01 | APL bioengineering |
| 34291416 | Fragile X-Related Protein 1 (FXR1) Promotes Trophoblast Migration at Early Pregnancy via Downregulation of GDF-15 Expression. | Wei Hong | 2022-01-01 | Reproductive sciences (Thousand Oaks, Calif.) |
| 35294905 | The RNA-binding protein fragile-X mental retardation autosomal 1 (FXR1) modulates glioma cells sensitivity to temozolomide by regulating ferroptosis. | Yiting Wei | 2022-05-07 | Biochemical and biophysical research communications |
| 30687136 | Antipsychotic Drug Responsiveness and Dopamine Receptor Signaling; Old Players and New Prospects. | Antonio Rampino | 2018-01-01 | Frontiers in psychiatry |
| 32995776 | Genes with 5' terminal oligopyrimidine tracts preferentially escape global suppression of translation by the SARS-CoV-2 Nsp1 protein. | Shilpa Rao | 2021-05-25 | bioRxiv : the preprint server for biology |
| 27229378 | Regulation of monocyte induced cell migration by the RNA binding protein, FXR1. | O O Le | 2016-07-17 | Cell cycle (Georgetown, Tex.) |
| 23582851 | Fragile hearts: new insights into translational control in cardiac muscle. | C Daniela Zarnescu | 2013-11-01 | Trends in cardiovascular medicine |
| 35887344 | Amyloid Properties of the FXR1 Protein Are Conserved in Evolution of Vertebrates. | E Maria Velizhanina | 2022-07-20 | International journal of molecular sciences |
| 27606879 | RNA-Binding Protein FXR1 Regulates p21 and TERC RNA to Bypass p53-Mediated Cellular Senescence in OSCC. | Mrinmoyee Majumder | 2016-09-01 | PLoS genetics |
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| FXR1-208 | ENST00000469882 | 570 | 119aa | ENSP00000419793 |
| FXR1-218 | ENST00000484790 | 571 | 75aa | ENSP00000417125 |
| FXR1-206 | ENST00000465551 | 572 | 94aa | ENSP00000418724 |
| FXR1-201 | ENST00000305586 | 3277 | 536aa | ENSP00000307633 |
| FXR1-211 | ENST00000475315 | 2341 | 54aa | ENSP00000513935 |
| FXR1-221 | ENST00000491674 | 365 | 19aa | ENSP00000419047 |
| FXR1-223 | ENST00000698793 | 5294 | 568aa | ENSP00000513936 |
| FXR1-224 | ENST00000698794 | 3593 | 650aa | ENSP00000513937 |
| FXR1-202 | ENST00000357559 | 8343 | 621aa | ENSP00000350170 |
| FXR1-213 | ENST00000479176 | 541 | No protein | - |
| FXR1-220 | ENST00000491062 | 1768 | 490aa | ENSP00000420643 |
| FXR1-207 | ENST00000468861 | 1989 | 454aa | ENSP00000420515 |
| FXR1-203 | ENST00000445140 | 2651 | 539aa | ENSP00000388828 |
| FXR1-209 | ENST00000472339 | 2130 | No protein | - |
| FXR1-219 | ENST00000484958 | 824 | 123aa | ENSP00000419933 |
| FXR1-214 | ENST00000480918 | 1996 | 608aa | ENSP00000418097 |
| FXR1-217 | ENST00000484042 | 562 | 151aa | ENSP00000417513 |
| FXR1-205 | ENST00000461801 | 410 | No protein | - |
| FXR1-210 | ENST00000473375 | 483 | No protein | - |
| FXR1-204 | ENST00000461044 | 601 | No protein | - |
| FXR1-222 | ENST00000498658 | 683 | No protein | - |
| FXR1-212 | ENST00000476672 | 976 | No protein | - |
| FXR1-216 | ENST00000482125 | 739 | 246aa | ENSP00000418027 |
| FXR1-215 | ENST00000481383 | 909 | No protein | - |
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
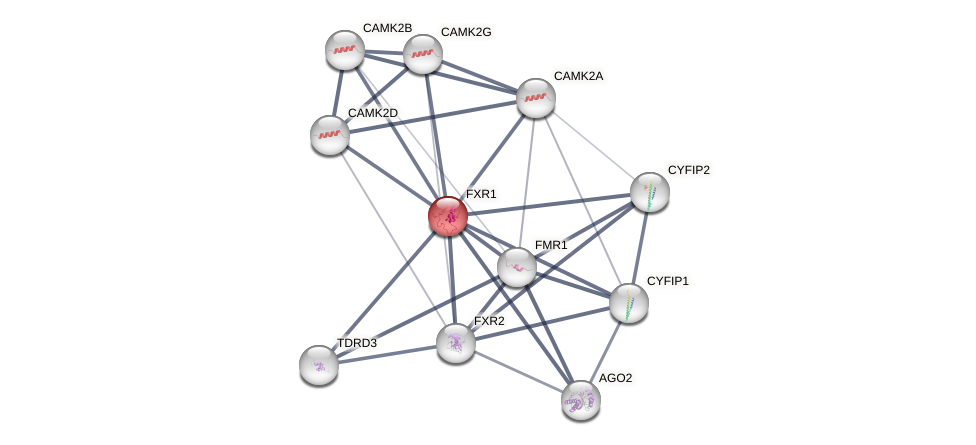
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000114416 | homo_sapiens | ENSP00000359506 | 66.6139 | 57.1203 | homo_sapiens | ENSP00000350170 | 67.7939 | 58.132 |
| ENSG00000114416 | homo_sapiens | ENSP00000250113 | 69.688 | 59.2868 | homo_sapiens | ENSP00000350170 | 75.5233 | 64.2512 |
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 97.4235 | 97.2625 | nomascus_leucogenys | ENSNLEP00000028687 | 91.528 | 91.3767 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 100 | 100 | pan_paniscus | ENSPPAP00000029899 | 79.4118 | 79.4118 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 100 | 100 | pongo_abelii | ENSPPYP00000031456 | 95.5385 | 95.5385 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 100 | 100 | pan_troglodytes | ENSPTRP00000026942 | 79.4118 | 79.4118 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 100 | 100 | gorilla_gorilla | ENSGGOP00000035407 | 79.8201 | 79.8201 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 97.9066 | 96.2963 | ornithorhynchus_anatinus | ENSOANP00000039068 | 81.5013 | 80.1609 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 98.3897 | 97.1014 | sarcophilus_harrisii | ENSSHAP00000038077 | 89.9853 | 88.8071 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 72.9469 | 71.9807 | notamacropus_eugenii | ENSMEUP00000004304 | 75 | 74.0066 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 64.8953 | 64.4122 | choloepus_hoffmanni | ENSCHOP00000011407 | 86.4807 | 85.8369 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 63.124 | 56.6828 | dasypus_novemcinctus | ENSDNOP00000018758 | 72.8625 | 65.4275 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 100 | 99.6779 | dasypus_novemcinctus | ENSDNOP00000014787 | 95.8333 | 95.5247 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 99.3559 | 98.0676 | erinaceus_europaeus | ENSEEUP00000006255 | 99.3559 | 98.0676 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 89.211 | 88.5668 | echinops_telfairi | ENSETEP00000005588 | 98.4014 | 97.6909 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 99.839 | 99.5169 | callithrix_jacchus | ENSCJAP00000023572 | 95.679 | 95.3704 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 100 | 100 | cercocebus_atys | ENSCATP00000036252 | 95.8333 | 95.8333 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 100 | 100 | macaca_fascicularis | ENSMFAP00000001144 | 78.0151 | 78.0151 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 100 | 100 | macaca_mulatta | ENSMMUP00000038773 | 77.8195 | 77.8195 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 100 | 100 | macaca_nemestrina | ENSMNEP00000015283 | 95.8333 | 95.8333 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 100 | 100 | papio_anubis | ENSPANP00000024003 | 77.8195 | 77.8195 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 97.4235 | 97.2625 | mandrillus_leucophaeus | ENSMLEP00000034500 | 91.528 | 91.3767 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 99.839 | 99.3559 | tursiops_truncatus | ENSTTRP00000013937 | 100 | 99.5161 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 100 | 99.6779 | vulpes_vulpes | ENSVVUP00000008910 | 95.8333 | 95.5247 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 99.1948 | 98.2287 | mus_spretus | MGP_SPRETEiJ_P0059563 | 90.9897 | 90.1034 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 100 | 99.5169 | equus_asinus | ENSEASP00005043193 | 91.7282 | 91.2851 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 100 | 99.5169 | capra_hircus | ENSCHIP00000017285 | 95.8333 | 95.3704 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 99.5169 | 98.8728 | loxodonta_africana | ENSLAFP00000029244 | 91.2851 | 90.6942 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 86.1514 | 85.8293 | panthera_leo | ENSPLOP00000030656 | 99.2579 | 98.8868 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 100 | 99.5169 | panthera_leo | ENSPLOP00000024354 | 100 | 99.5169 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 100 | 99.6779 | ailuropoda_melanoleuca | ENSAMEP00000042117 | 95.8333 | 95.5247 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 100 | 99.5169 | bos_indicus_hybrid | ENSBIXP00005014564 | 91.7282 | 91.2851 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 99.839 | 99.1948 | oryctolagus_cuniculus | ENSOCUP00000042126 | 79.3854 | 78.8732 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 100 | 99.5169 | equus_caballus | ENSECAP00000023539 | 88.9685 | 88.5387 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 100 | 99.6779 | canis_lupus_familiaris | ENSCAFP00845027124 | 95.8333 | 95.5247 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 100 | 99.5169 | panthera_pardus | ENSPPRP00000010790 | 100 | 99.5169 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 100 | 99.1948 | marmota_marmota_marmota | ENSMMMP00000003770 | 91.7282 | 90.9897 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 99.6779 | 99.3559 | balaenoptera_musculus | ENSBMSP00010006590 | 100 | 99.6769 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 81.6425 | 81.3204 | balaenoptera_musculus | ENSBMSP00010018070 | 100 | 99.6055 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 99.3559 | 99.0338 | cavia_porcellus | ENSCPOP00000024292 | 91.1374 | 90.8419 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 99.839 | 99.3559 | felis_catus | ENSFCAP00000060289 | 100 | 99.5161 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 86.3124 | 85.9903 | ursus_americanus | ENSUAMP00000007975 | 100 | 99.6269 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 95.9742 | 94.847 | monodelphis_domestica | ENSMODP00000026620 | 90.8537 | 89.7866 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 100 | 99.5169 | bison_bison_bison | ENSBBBP00000007818 | 100 | 99.5169 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 100 | 99.6779 | monodon_monoceros | ENSMMNP00015001208 | 100 | 99.6779 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 99.6779 | 99.3559 | sus_scrofa | ENSSSCP00000023407 | 91.4328 | 91.1374 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 82.9308 | 81.1594 | microcebus_murinus | ENSMICP00000030525 | 95.9032 | 93.8548 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 94.847 | 94.525 | microcebus_murinus | ENSMICP00000051455 | 91.0355 | 90.7264 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 68.1159 | 63.124 | octodon_degus | ENSODEP00000001258 | 84.2629 | 78.0876 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 99.0338 | 98.7118 | octodon_degus | ENSODEP00000001355 | 90.8419 | 90.5465 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 78.2609 | 77.1337 | octodon_degus | ENSODEP00000008287 | 91.0112 | 89.7004 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 94.0419 | 93.7198 | jaculus_jaculus | ENSJJAP00000017724 | 90.5426 | 90.2326 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 93.3977 | 92.9147 | ovis_aries_rambouillet | ENSOARP00020007735 | 89.2308 | 88.7692 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 86.3124 | 85.9903 | ursus_maritimus | ENSUMAP00000009345 | 100 | 99.6269 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 91.6264 | 90.6602 | ochotona_princeps | ENSOPRP00000008446 | 94.5183 | 93.5216 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 100 | 99.6779 | delphinapterus_leucas | ENSDLEP00000016254 | 100 | 99.6779 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 100 | 99.6779 | physeter_catodon | ENSPCTP00005005510 | 92.6866 | 92.3881 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 99.1948 | 98.2287 | mus_caroli | MGP_CAROLIEiJ_P0057306 | 90.9897 | 90.1034 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 99.1948 | 98.0676 | mus_musculus | ENSMUSP00000001620 | 90.9897 | 89.9557 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 85.5072 | 84.219 | dipodomys_ordii | ENSDORP00000024148 | 88.3527 | 87.0216 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 100 | 99.6779 | mustela_putorius_furo | ENSMPUP00000002309 | 95.8333 | 95.5247 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 99.5169 | 99.3559 | aotus_nancymaae | ENSANAP00000016316 | 88.412 | 88.269 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 94.0419 | 93.3977 | saimiri_boliviensis_boliviensis | ENSSBOP00000035058 | 89.7081 | 89.0937 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 97.1014 | 96.7794 | vicugna_pacos | ENSVPAP00000003128 | 100 | 99.6683 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 100 | 100 | chlorocebus_sabaeus | ENSCSAP00000009348 | 95.8333 | 95.8333 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 55.7166 | 55.5556 | tupaia_belangeri | ENSTBEP00000006373 | 64.9156 | 64.728 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 99.1948 | 98.3897 | mesocricetus_auratus | ENSMAUP00000001655 | 90.9897 | 90.2511 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 99.839 | 99.3559 | phocoena_sinus | ENSPSNP00000002327 | 97.7918 | 97.3186 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 100 | 99.6779 | camelus_dromedarius | ENSCDRP00005002096 | 95.8333 | 95.5247 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 97.4235 | 96.7794 | carlito_syrichta | ENSTSYP00000021527 | 91.3897 | 90.7855 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 100 | 99.5169 | panthera_tigris_altaica | ENSPTIP00000023281 | 100 | 99.5169 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 99.3559 | 98.5507 | rattus_norvegicus | ENSRNOP00000095711 | 91.1374 | 90.3988 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 92.1095 | 91.9485 | prolemur_simus | ENSPSMP00000016668 | 91.9614 | 91.8006 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 97.5845 | 96.2963 | microtus_ochrogaster | ENSMOCP00000019859 | 88.9868 | 87.812 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 76.4895 | 76.3285 | sorex_araneus | ENSSARP00000010816 | 78.6424 | 78.4768 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 96.4573 | 95.8132 | peromyscus_maniculatus_bairdii | ENSPEMP00000012759 | 88.4786 | 87.8877 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 98.5507 | 98.0676 | bos_mutus | ENSBMUP00000023337 | 100 | 99.5098 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 99.3559 | 98.3897 | mus_spicilegus | ENSMSIP00000013782 | 91.1374 | 90.2511 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 99.1948 | 98.3897 | cricetulus_griseus_chok1gshd | ENSCGRP00001027943 | 95.0617 | 94.2901 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 84.5411 | 84.38 | pteropus_vampyrus | ENSPVAP00000002668 | 92.9204 | 92.7434 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 97.4235 | 96.9404 | myotis_lucifugus | ENSMLUP00000006253 | 91.528 | 91.0741 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 98.2287 | 97.1014 | vombatus_ursinus | ENSVURP00010015692 | 97.9133 | 96.7897 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 99.839 | 99.3559 | rhinolophus_ferrumequinum | ENSRFEP00010032351 | 95.679 | 95.216 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 100 | 99.5169 | neovison_vison | ENSNVIP00000021418 | 100 | 99.5169 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 100 | 99.5169 | bos_taurus | ENSBTAP00000068817 | 95.8333 | 95.3704 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 99.5169 | 99.0338 | heterocephalus_glaber_female | ENSHGLP00000026444 | 91.2851 | 90.8419 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 89.8551 | 89.533 | rhinopithecus_bieti | ENSRBIP00000026194 | 94.898 | 94.5578 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 100 | 99.6779 | canis_lupus_dingo | ENSCAFP00020030178 | 100 | 99.6779 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 100 | 99.1948 | sciurus_vulgaris | ENSSVLP00005007206 | 100 | 99.1948 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 94.3639 | 93.3977 | phascolarctos_cinereus | ENSPCIP00000035593 | 92.575 | 91.6272 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 47.9871 | 46.8599 | procavia_capensis | ENSPCAP00000013918 | 49.3377 | 48.1788 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 97.7456 | 95.9742 | nannospalax_galili | ENSNGAP00000023071 | 90.0593 | 88.4273 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 100 | 99.5169 | bos_grunniens | ENSBGRP00000003331 | 95.8333 | 95.3704 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 99.0338 | 98.8728 | chinchilla_lanigera | ENSCLAP00000020701 | 90.9763 | 90.8284 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 100 | 99.5169 | cervus_hanglu_yarkandensis | ENSCHYP00000023931 | 95.8333 | 95.3704 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 99.6779 | 99.3559 | catagonus_wagneri | ENSCWAP00000014313 | 99.6779 | 99.3559 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 100 | 99.1948 | ictidomys_tridecemlineatus | ENSSTOP00000018711 | 91.7282 | 90.9897 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 100 | 99.6779 | otolemur_garnettii | ENSOGAP00000007847 | 100 | 99.6779 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 92.4316 | 91.3043 | cebus_imitator | ENSCCAP00000014582 | 88.8545 | 87.7709 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 100 | 99.1948 | urocitellus_parryii | ENSUPAP00010028597 | 95.8333 | 95.0617 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 72.4638 | 71.9807 | moschus_moschiferus | ENSMMSP00000022985 | 99.1189 | 98.4582 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 100 | 99.5169 | moschus_moschiferus | ENSMMSP00000001937 | 100 | 99.5169 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 100 | 100 | rhinopithecus_roxellana | ENSRROP00000021391 | 95.8333 | 95.8333 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 99.3559 | 98.3897 | mus_pahari | MGP_PahariEiJ_P0068294 | 91.1374 | 90.2511 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 94.525 | 93.7198 | propithecus_coquereli | ENSPCOP00000016613 | 89.2097 | 88.4498 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 57.649 | 40.9018 | drosophila_melanogaster | FBpp0300445 | 49.1084 | 34.8423 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 47.9871 | 35.2657 | ciona_intestinalis | ENSCINP00000017388 | 63.6752 | 46.7949 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 85.6683 | 77.9388 | callorhinchus_milii | ENSCMIP00000032834 | 77.7778 | 70.7602 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 91.4654 | 83.5749 | latimeria_chalumnae | ENSLACP00000012821 | 83.6524 | 76.4359 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 89.8551 | 82.4477 | lepisosteus_oculatus | ENSLOCP00000000652 | 81.6984 | 74.9634 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 87.4396 | 78.0998 | clupea_harengus | ENSCHAP00000043182 | 74.8966 | 66.8966 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 83.0918 | 72.6248 | danio_rerio | ENSDARP00000124192 | 80 | 69.9225 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 86.6345 | 75.5233 | carassius_auratus | ENSCARP00000104317 | 82.1374 | 71.6031 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 86.6345 | 76.3285 | carassius_auratus | ENSCARP00000033621 | 80.7808 | 71.1712 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 85.9903 | 76.1675 | astyanax_mexicanus | ENSAMXP00000054329 | 79.5827 | 70.4918 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 81.9646 | 70.3704 | ictalurus_punctatus | ENSIPUP00000036726 | 76.5414 | 65.7143 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 79.3881 | 67.7939 | esox_lucius | ENSELUP00000073292 | 74.4713 | 63.5952 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 84.058 | 73.1079 | electrophorus_electricus | ENSEEEP00000035804 | 77.3333 | 67.2593 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 60.0644 | 50.2415 | oncorhynchus_kisutch | ENSOKIP00005103906 | 77.8706 | 65.1357 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 84.5411 | 73.913 | oncorhynchus_kisutch | ENSOKIP00005079201 | 77.548 | 67.7991 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 80.0322 | 69.5652 | oncorhynchus_mykiss | ENSOMYP00000032527 | 76.935 | 66.8731 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 74.2351 | 65.2174 | oncorhynchus_mykiss | ENSOMYP00000056713 | 80.5944 | 70.8042 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 54.4283 | 48.1481 | oncorhynchus_mykiss | ENSOMYP00000125931 | 72.6882 | 64.3011 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 48.7923 | 43.8003 | oncorhynchus_mykiss | ENSOMYP00000115826 | 78.4974 | 70.4663 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 84.058 | 73.752 | salmo_salar | ENSSSAP00000127651 | 77.3333 | 67.8519 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 21.5781 | 17.7134 | salmo_salar | ENSSSAP00000175571 | 67.3367 | 55.2764 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 79.7101 | 70.0483 | salmo_trutta | ENSSTUP00000090548 | 76.1538 | 66.9231 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 72.3027 | 62.4799 | gadus_morhua | ENSGMOP00000041282 | 67.8248 | 58.6103 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 32.2061 | 28.3414 | fundulus_heteroclitus | ENSFHEP00000002889 | 77.2201 | 67.9537 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 25.7649 | 22.2222 | fundulus_heteroclitus | ENSFHEP00000002902 | 86.4865 | 74.5946 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 75.0403 | 65.5395 | poecilia_reticulata | ENSPREP00000033555 | 81.6112 | 71.2785 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 83.5749 | 71.9807 | xiphophorus_maculatus | ENSXMAP00000000684 | 79.9692 | 68.8752 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 82.4477 | 71.8196 | oryzias_latipes | ENSORLP00000014777 | 75.405 | 65.6848 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 75.0403 | 65.2174 | cyclopterus_lumpus | ENSCLMP00005033573 | 74.9196 | 65.1125 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 83.8969 | 71.9807 | oreochromis_niloticus | ENSONIP00000039382 | 70.9809 | 60.8992 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 84.058 | 72.1417 | haplochromis_burtoni | ENSHBUP00000003599 | 71.1172 | 61.0354 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 84.058 | 71.8196 | astatotilapia_calliptera | ENSACLP00000038250 | 73.7288 | 62.9944 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 84.219 | 71.9807 | sparus_aurata | ENSSAUP00010024367 | 71.5458 | 61.1491 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 84.38 | 72.7858 | lates_calcarifer | ENSLCAP00010005172 | 70.9066 | 61.1637 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 93.5588 | 88.0837 | xenopus_tropicalis | ENSXETP00000042154 | 86.2018 | 81.1573 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 97.4235 | 95.3301 | chrysemys_picta_bellii | ENSCPBP00000000236 | 89.233 | 87.3156 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 82.2866 | 80.0322 | sphenodon_punctatus | ENSSPUP00000018836 | 96.2335 | 93.597 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 92.2705 | 88.5668 | crocodylus_porosus | ENSCPRP00005015015 | 85.2679 | 81.8452 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 94.2029 | 89.533 | laticauda_laticaudata | ENSLLTP00000021625 | 85.9031 | 81.6446 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 95.3301 | 91.4654 | notechis_scutatus | ENSNSUP00000005698 | 91.0769 | 87.3846 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 95.4911 | 91.6264 | pseudonaja_textilis | ENSPTXP00000025330 | 87.5923 | 84.0473 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 96.7794 | 93.3977 | anas_platyrhynchos_platyrhynchos | ENSAPLP00000028519 | 92.4615 | 89.2308 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 96.2963 | 92.7536 | gallus_gallus | ENSGALP00010023299 | 88.3309 | 85.0812 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 93.2367 | 88.8889 | meleagris_gallopavo | ENSMGAP00000009956 | 82.1277 | 78.2979 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 91.3043 | 87.6006 | serinus_canaria | ENSSCAP00000017190 | 87.0968 | 83.5638 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 96.6184 | 92.9147 | parus_major | ENSPMJP00000002716 | 81.3008 | 78.1843 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 92.9147 | 89.533 | anolis_carolinensis | ENSACAP00000003925 | 86.7669 | 83.609 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 75.3623 | 66.1836 | neolamprologus_brichardi | ENSNBRP00000027638 | 77.6119 | 68.1592 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 94.2029 | 90.3382 | naja_naja | ENSNNAP00000015282 | 86.4106 | 82.8656 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 95.3301 | 92.2705 | podarcis_muralis | ENSPMRP00000006193 | 87.3156 | 84.5133 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 91.9485 | 87.9227 | geospiza_fortis | ENSGFOP00000010374 | 87.3089 | 83.4862 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 96.4573 | 92.9147 | taeniopygia_guttata | ENSTGUP00000010912 | 88.4786 | 85.229 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 67.6329 | 60.7085 | erpetoichthys_calabaricus | ENSECRP00000018660 | 84.6774 | 76.0081 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 80.0322 | 68.1159 | kryptolebias_marmoratus | ENSKMAP00000006217 | 75.9939 | 64.6789 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 95.8132 | 92.1095 | salvator_merianae | ENSSMRP00000030785 | 87.7581 | 84.3658 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 83.7359 | 71.9807 | oryzias_javanicus | ENSOJAP00000045332 | 76.1347 | 65.4466 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 76.4895 | 66.1836 | amphilophus_citrinellus | ENSACIP00000030177 | 80.5085 | 69.661 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 76.9726 | 66.3446 | dicentrarchus_labrax | ENSDLAP00005070178 | 74.2236 | 63.9752 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 80.5153 | 70.2093 | amphiprion_percula | ENSAPEP00000021047 | 79.6178 | 69.4268 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 82.7697 | 71.8196 | oryzias_sinensis | ENSOSIP00000028560 | 75.9232 | 65.8789 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 93.3977 | 90.6602 | pelodiscus_sinensis | ENSPSIP00000008435 | 86.3095 | 83.7798 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 97.2625 | 95.1691 | gopherus_evgoodei | ENSGEVP00005010414 | 92.7803 | 90.7834 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 93.8808 | 90.9823 | chelonoidis_abingdonii | ENSCABP00000026720 | 86.756 | 84.0774 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 85.3462 | 73.2689 | seriola_dumerili | ENSSDUP00000022800 | 74.4382 | 63.9045 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 80.1932 | 68.277 | cottoperca_gobio | ENSCGOP00000004246 | 73.8872 | 62.908 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 90.4992 | 86.7955 | struthio_camelus_australis | ENSSCUP00000023769 | 87.5389 | 83.9564 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 96.7794 | 93.3977 | anser_brachyrhynchus | ENSABRP00000004998 | 92.4615 | 89.2308 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 81.1594 | 69.4042 | gasterosteus_aculeatus | ENSGACP00000008025 | 75 | 64.1369 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 85.3462 | 72.7858 | cynoglossus_semilaevis | ENSCSEP00000002405 | 75.1773 | 64.1135 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 84.058 | 72.7858 | poecilia_formosa | ENSPFOP00000014815 | 77.2189 | 66.8639 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 85.5072 | 73.2689 | myripristis_murdjan | ENSMMDP00005041136 | 78.4343 | 67.2083 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 83.0918 | 71.8196 | sander_lucioperca | ENSSLUP00000021572 | 67.3629 | 58.2245 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 96.6184 | 93.2367 | strigops_habroptila | ENSSHBP00005013879 | 88.6263 | 85.5244 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 79.5491 | 68.7601 | amphiprion_ocellaris | ENSAOCP00000030871 | 82.6087 | 71.4047 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 97.4235 | 95.3301 | terrapene_carolina_triunguis | ENSTMTP00000013782 | 92.9339 | 90.937 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 84.38 | 74.0741 | hucho_hucho | ENSHHUP00000055964 | 77.4003 | 67.9468 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 49.9195 | 42.351 | hucho_hucho | ENSHHUP00000086506 | 79.6915 | 67.6093 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 82.2866 | 71.1755 | betta_splendens | ENSBSLP00000014686 | 71.669 | 61.9916 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 85.8293 | 75.3623 | pygocentrus_nattereri | ENSPNAP00000015162 | 77.6968 | 68.2216 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 83.7359 | 71.6586 | anabas_testudineus | ENSATEP00000039105 | 70.1754 | 60.054 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 35.1047 | 24.3156 | ciona_savignyi | ENSCSAVP00000012645 | 66.8712 | 46.319 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 80.5153 | 70.5314 | seriola_lalandi_dorsalis | ENSSLDP00000023572 | 82.7815 | 72.5166 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 79.8712 | 67.7939 | labrus_bergylta | ENSLBEP00000015268 | 70.9585 | 60.2289 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 84.219 | 71.9807 | labrus_bergylta | ENSLBEP00000019601 | 70.3903 | 60.1615 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 83.8969 | 71.8196 | pundamilia_nyererei | ENSPNYP00000018611 | 75.5072 | 64.6377 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 85.3462 | 73.913 | mastacembelus_armatus | ENSMAMP00000039532 | 76.5896 | 66.3295 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 86.1514 | 73.591 | stegastes_partitus | ENSSPAP00000013976 | 77.2006 | 65.9452 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 37.1981 | 32.2061 | oncorhynchus_tshawytscha | ENSOTSP00005065102 | 89.1892 | 77.2201 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 59.4203 | 51.6908 | oncorhynchus_tshawytscha | ENSOTSP00005047867 | 77.6842 | 67.5789 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 33.4944 | 29.7907 | oncorhynchus_tshawytscha | ENSOTSP00005065106 | 81.8898 | 72.8346 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 85.5072 | 73.1079 | acanthochromis_polyacanthus | ENSAPOP00000014145 | 76.8452 | 65.7019 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 96.7794 | 93.3977 | aquila_chrysaetos_chrysaetos | ENSACCP00020001475 | 92.4615 | 89.2308 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 84.219 | 71.1755 | nothobranchius_furzeri | ENSNFUP00015034857 | 48.3811 | 40.8881 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 84.38 | 72.4638 | cyprinodon_variegatus | ENSCVAP00000001692 | 74.3262 | 63.8298 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 86.1514 | 75.0403 | denticeps_clupeoides | ENSDCDP00000063581 | 78.1022 | 68.0292 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 84.5411 | 72.7858 | larimichthys_crocea | ENSLCRP00005036723 | 74.4681 | 64.1135 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 81.3204 | 68.438 | takifugu_rubripes | ENSTRUP00000060722 | 73.6152 | 61.9534 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 94.847 | 91.1433 | ficedula_albicollis | ENSFALP00000008651 | 84.505 | 81.2052 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 69.7262 | 60.3865 | hippocampus_comes | ENSHCOP00000008122 | 76.9094 | 66.6075 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 39.7746 | 33.8164 | hippocampus_comes | ENSHCOP00000022454 | 87.9004 | 74.7331 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 57.971 | 50.0805 | hippocampus_comes | ENSHCOP00000022481 | 77.2532 | 66.7382 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 86.6345 | 75.6844 | cyprinus_carpio_carpio | ENSCCRP00000092527 | 81.3918 | 71.1044 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 86.7955 | 76.6506 | cyprinus_carpio_carpio | ENSCCRP00000177535 | 83.8258 | 74.028 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 96.4573 | 92.7536 | coturnix_japonica | ENSCJPP00005017450 | 92.1538 | 88.6154 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 83.8969 | 72.6248 | poecilia_latipinna | ENSPLAP00000031304 | 77.1852 | 66.8148 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 83.7359 | 73.913 | sinocyclocheilus_grahami | ENSSGRP00000103722 | 79.8771 | 70.5069 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 83.8969 | 74.3961 | sinocyclocheilus_grahami | ENSSGRP00000016087 | 79.542 | 70.5343 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 83.8969 | 71.8196 | maylandia_zebra | ENSMZEP00005020905 | 75.7267 | 64.8256 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 80.6763 | 67.6329 | tetraodon_nigroviridis | ENSTNIP00000006390 | 73.5683 | 61.674 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 15.62 | 9.82287 | tetraodon_nigroviridis | ENSTNIP00000009608 | 62.1795 | 39.1026 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 86.6345 | 75.6844 | paramormyrops_kingsleyae | ENSPKIP00000015252 | 78.655 | 68.7134 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 77.4557 | 65.0564 | paramormyrops_kingsleyae | ENSPKIP00000034996 | 74.3431 | 62.442 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 74.8792 | 63.124 | scleropages_formosus | ENSSFOP00015057045 | 72.093 | 60.7752 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 87.1176 | 78.4219 | scleropages_formosus | ENSSFOP00015078683 | 79.2094 | 71.3031 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 90.1771 | 83.8969 | leptobrachium_leishanense | ENSLLEP00000047120 | 82.5959 | 76.8437 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 61.3527 | 58.6151 | leptobrachium_leishanense | ENSLLEP00000038952 | 85.618 | 81.7978 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 21.9002 | 19.6457 | leptobrachium_leishanense | ENSLLEP00000006404 | 73.913 | 66.3043 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 84.058 | 72.1417 | scophthalmus_maximus | ENSSMAP00000031415 | 70.0671 | 60.1342 |
| ENSG00000114416 | homo_sapiens | ENSP00000350170 | 83.0918 | 71.6586 | oryzias_melastigma | ENSOMEP00000028582 | 75.549 | 65.1537 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0000381 | involved_in regulation of alternative mRNA splicing, via spliceosome | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0001934 | involved_in positive regulation of protein phosphorylation | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0003723 | enables RNA binding | 2 | HDA,IDA | Homo_sapiens(9606) | Function |
| GO:0003730 | enables mRNA 3-UTR binding | 2 | IBA,IDA | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005634 | is_active_in nucleus | 1 | IBA | Homo_sapiens(9606) | Component |
| GO:0005635 | is_active_in nuclear envelope | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005730 | located_in nucleolus | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0005737 | is_active_in cytoplasm | 2 | IDA,IMP | Homo_sapiens(9606) | Component |
| GO:0005829 | located_in cytosol | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005844 | part_of polysome | 1 | IBA | Homo_sapiens(9606) | Component |
| GO:0006915 | involved_in apoptotic process | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0007286 | involved_in spermatid development | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0007517 | involved_in muscle organ development | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0010494 | is_active_in cytoplasmic stress granule | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0014069 | is_active_in postsynaptic density | 1 | IBA | Homo_sapiens(9606) | Component |
| GO:0016020 | located_in membrane | 2 | HDA,IBA | Homo_sapiens(9606) | Component |
| GO:0017148 | involved_in negative regulation of translation | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0021542 | involved_in dentate gyrus development | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0030424 | is_active_in axon | 1 | IBA | Homo_sapiens(9606) | Component |
| GO:0030426 | is_active_in growth cone | 1 | IBA | Homo_sapiens(9606) | Component |
| GO:0032720 | involved_in negative regulation of tumor necrosis factor production | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0033592 | enables RNA strand annealing activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0035925 | enables mRNA 3-UTR AU-rich region binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0036464 | is_active_in cytoplasmic ribonucleoprotein granule | 2 | IBA,IDA | Homo_sapiens(9606) | Component |
| GO:0042803 | enables protein homodimerization activity | 2 | IBA,IDA | Homo_sapiens(9606) | Function |
| GO:0043025 | is_active_in neuronal cell body | 1 | IBA | Homo_sapiens(9606) | Component |
| GO:0043034 | located_in costamere | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0043197 | is_active_in dendritic spine | 1 | IBA | Homo_sapiens(9606) | Component |
| GO:0043232 | is_active_in intracellular non-membrane-bounded organelle | 1 | ISS | Homo_sapiens(9606) | Component |
| GO:0043488 | involved_in regulation of mRNA stability | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0044326 | is_active_in dendritic spine neck | 1 | IBA | Homo_sapiens(9606) | Component |
| GO:0045182 | enables translation regulator activity | 1 | IBA | Homo_sapiens(9606) | Function |
| GO:0045187 | involved_in regulation of circadian sleep/wake cycle, sleep | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0045727 | involved_in positive regulation of translation | 2 | IBA,IDA | Homo_sapiens(9606) | Process |
| GO:0046982 | enables protein heterodimerization activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0048471 | located_in perinuclear region of cytoplasm | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0050728 | involved_in negative regulation of inflammatory response | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0050767 | involved_in regulation of neurogenesis | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0051292 | involved_in nuclear pore complex assembly | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0051489 | involved_in regulation of filopodium assembly | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0051664 | involved_in nuclear pore localization | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0051966 | involved_in regulation of synaptic transmission, glutamatergic | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0060538 | involved_in skeletal muscle organ development | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0061157 | involved_in mRNA destabilization | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0098793 | is_active_in presynapse | 1 | IBA | Homo_sapiens(9606) | Component |
| GO:0098978 | located_in glutamatergic synapse | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0140693 | enables molecular condensate scaffold activity | 1 | ISS | Homo_sapiens(9606) | Function |
| GO:0140694 | involved_in non-membrane-bounded organelle assembly | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:1900272 | involved_in negative regulation of long-term synaptic potentiation | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:1902737 | is_active_in dendritic filopodium | 1 | IBA | Homo_sapiens(9606) | Component |
| GO:2000637 | involved_in positive regulation of miRNA-mediated gene silencing | 1 | IDA | Homo_sapiens(9606) | Process |