Welcome to
RBP World!
Functions and Diseases
| RBP Type | Canonical_RBPs |
| Diseases | CancerCHIKVSARS-COV-OC43FlavivirusesHIVRVSARS-COV-2SINVDengueZika |
| Drug | N.A. |
| Main interacting RNAs | mRNA |
| Moonlighting functions | N.A. |
| Localizations | Stress granucleP-body |
| BulkPerturb-seq | DataSet_01_104 DataSet_01_414 |
Description
| Ensembl ID | ENSG00000099783 | Gene ID | 4670 | Accession | 5046 |
| Symbol | HNRNPM | Alias | CEAR;HNRPM;HTGR1;NAGR1;HNRPM4;HNRNPM4;hnRNPM | Full Name | heterogeneous nuclear ribonucleoprotein M |
| Status | Confidence | Length | 44348 bases | Strand | Plus strand |
| Position | 19 : 8444767 - 8489114 | RNA binding domain | RRM_1 , HnRNP_M_NLS | ||
| Summary | This gene belongs to the subfamily of ubiquitously expressed heterogeneous nuclear ribonucleoproteins (hnRNPs). The hnRNPs are RNA binding proteins and they complex with heterogeneous nuclear RNA (hnRNA). These proteins are associated with pre-mRNAs in the nucleus and appear to influence pre-mRNA processing and other aspects of mRNA metabolism and transport. While all of the hnRNPs are present in the nucleus, some seem to shuttle between the nucleus and the cytoplasm. The hnRNP proteins have distinct nucleic acid binding properties. The protein encoded by this gene has three repeats of quasi-RRM domains that bind to RNAs. This protein also constitutes a monomer of the N-acetylglucosamine-specific receptor which is postulated to trigger selective recycling of immature GlcNAc-bearing thyroglobulin molecules. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Aug 2011] | ||||
RNA binding domains (RBDs)
| Protein | Domain | Pfam ID | E-value | Domain number |
|---|---|---|---|---|
| ENSP00000325732 | RRM_1 | PF00076 | 4.8e-52 | 1 |
| ENSP00000325732 | RRM_1 | PF00076 | 4.8e-52 | 2 |
| ENSP00000325732 | RRM_1 | PF00076 | 4.8e-52 | 3 |
| ENSP00000325732 | HnRNP_M_NLS | PF11532 | 2.6e-17 | 4 |
| ENSP00000325376 | RRM_1 | PF00076 | 5.9e-52 | 1 |
| ENSP00000325376 | RRM_1 | PF00076 | 5.9e-52 | 2 |
| ENSP00000325376 | RRM_1 | PF00076 | 5.9e-52 | 3 |
| ENSP00000325376 | HnRNP_M_NLS | PF11532 | 2.8e-17 | 4 |
| ENSP00000483715 | RRM_1 | PF00076 | 2.4e-49 | 1 |
| ENSP00000483715 | RRM_1 | PF00076 | 2.4e-49 | 2 |
| ENSP00000483715 | RRM_1 | PF00076 | 2.4e-49 | 3 |
| ENSP00000483715 | HnRNP_M_NLS | PF11532 | 2.8e-17 | 4 |
| ENSP00000470932 | RRM_1 | PF00076 | 2e-31 | 1 |
| ENSP00000470932 | RRM_1 | PF00076 | 2e-31 | 2 |
| ENSP00000470932 | HnRNP_M_NLS | PF11532 | 9.9e-18 | 3 |
| ENSP00000472789 | RRM_1 | PF00076 | 2.1e-31 | 1 |
| ENSP00000472789 | RRM_1 | PF00076 | 2.1e-31 | 2 |
| ENSP00000472789 | HnRNP_M_NLS | PF11532 | 1e-17 | 3 |
| ENSP00000471339 | RRM_1 | PF00076 | 4.1e-29 | 1 |
| ENSP00000471339 | RRM_1 | PF00076 | 4.1e-29 | 2 |
| ENSP00000471339 | HnRNP_M_NLS | PF11532 | 7.9e-18 | 3 |
| ENSP00000470017 | RRM_1 | PF00076 | 8.9e-23 | 1 |
| ENSP00000470017 | HnRNP_M_NLS | PF11532 | 4.7e-18 | 2 |
| ENSP00000470635 | RRM_1 | PF00076 | 1.2e-09 | 1 |
| ENSP00000469681 | RRM_1 | PF00076 | 0.0003 | 1 |
| ENSP00000472599 | RRM_1 | PF00076 | 0.0011 | 1 |
| ENSP00000471530 | HnRNP_M_NLS | PF11532 | 3.2e-18 | 1 |
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|---|---|---|---|
| 30852162 | hnRNPM induces translation switch under hypoxia to promote colon cancer development. | Tsung-Ming Chen | 2019-03-01 | EBioMedicine |
| 31803364 | Tumor tissue hnRNP M and HSP 90α as potential predictors of disease-specific mortality in patients with early-stage cutaneous head and neck melanoma: A proteomics-based study. | Andro Košec | 2019-11-19 | Oncotarget |
| 32023846 | Poison-Exon Inclusion in DHX9 Reduces Its Expression and Sensitizes Ewing Sarcoma Cells to Chemotherapeutic Treatment. | Ramona Palombo | 2020-01-31 | Cells |
| 29675578 | The Parkinson's Disease-Linked Protein DJ-1 Associates with Cytoplasmic mRNP Granules During Stress and Neurodegeneration. | Mariaelena Repici | 2019-01-01 | Molecular neurobiology |
| 32811564 | RNA-recognition motif in Matrin-3 mediates neurodegeneration through interaction with hnRNPM. | Nandini Ramesh | 2020-08-18 | Acta neuropathologica communications |
| 28716093 | C. elegans SUP-46, an HNRNPM family RNA-binding protein that prevents paternally-mediated epigenetic sterility. | L Wendy Johnston | 2017-07-17 | BMC biology |
| 30813302 | The Relevance of Toxic AGEs (TAGE) Cytotoxicity to NASH Pathogenesis: A Mini-Review. | Akiko Sakasai-Sakai | 2019-02-22 | Nutrients |
| 36865202 | LINE-associated cryptic splicing induces dsRNA-mediated interferon response and tumor immunity. | Rong Zheng | 2023-02-24 | bioRxiv : the preprint server for biology |
| 33466816 | Alterations in Glucose Metabolism Due to Decreased Expression of Heterogeneous Nuclear Ribonucleoprotein M in Pancreatic Ductal Adenocarcinoma. | Jun-Ichi Takino | 2021-01-14 | Biology |
| 31980632 | The RNA-binding protein AKAP8 suppresses tumor metastasis by antagonizing EMT-associated alternative splicing. | Xiaohui Hu | 2020-01-24 | Nature communications |
| 37016304 | Differential splicing of neuronal genes in a Trem2*R47H mouse model mimics alterations associated with Alzheimer's disease. | S Ravi Pandey | 2023-04-04 | BMC genomics |
| 24840202 | Cell type-restricted activity of hnRNPM promotes breast cancer metastasis via regulating alternative splicing. | Yilin Xu | 2014-06-01 | Genes & development |
| 36417528 | Expression of expanded GGC repeats within NOTCH2NLC causes behavioral deficits and neurodegeneration in a mouse model of neuronal intranuclear inclusion disease. | Qiong Liu | 2022-11-25 | Science advances |
| 32732889 | Targeting QKI-7 in vivo restores endothelial cell function in diabetes. | Chunbo Yang | 2020-07-30 | Nature communications |
| 35732702 | SILAC kinase screen identifies potential MASTL substrates. | A Kamila Marzec | 2022-06-22 | Scientific reports |
| 30042172 | Coregulation of alternative splicing by hnRNPM and ESRP1 during EMT. | E Samuel Harvey | 2018-10-01 | RNA (New York, N.Y.) |
| 31737791 | Contribution of alternative splicing to breast cancer metastasis. | Xiangbing Meng | 2019-01-01 | Journal of cancer metastasis and treatment |
| 36978995 | Involvement of Intracellular TAGE and the TAGE-RAGE-ROS Axis in the Onset and Progression of NAFLD/NASH. | Akiko Sakasai-Sakai | 2023-03-19 | Antioxidants (Basel, Switzerland) |
| 36930678 | PCB: A pseudotemporal causality-based Bayesian approach to identify EMT-associated regulatory relationships of AS events and RBPs during breast cancer progression. | Liangjie Sun | 2023-03-01 | PLoS computational biology |
| 35883620 | Effects of Toxic AGEs (TAGE) on Human Health. | Masayoshi Takeuchi | 2022-07-12 | Cells |
| 36620748 | Epigenetic regulation of dental-derived stem cells and their application in pulp and periodontal regeneration. | Yuyang Chen | 2023-01-01 | PeerJ |
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| HNRNPM-202 | ENST00000348943 | 2568 | 691aa | ENSP00000325732 |
| HNRNPM-209 | ENST00000597813 | 630 | No protein | - |
| HNRNPM-213 | ENST00000600092 | 1086 | 352aa | ENSP00000470932 |
| HNRNPM-201 | ENST00000325495 | 2477 | 730aa | ENSP00000325376 |
| HNRNPM-214 | ENST00000600806 | 2373 | 99aa | ENSP00000471530 |
| HNRNPM-204 | ENST00000594907 | 1078 | 357aa | ENSP00000472789 |
| HNRNPM-206 | ENST00000596984 | 664 | 219aa | ENSP00000470017 |
| HNRNPM-215 | ENST00000601645 | 832 | 275aa | ENSP00000471339 |
| HNRNPM-203 | ENST00000593293 | 764 | No protein | - |
| HNRNPM-210 | ENST00000598367 | 298 | 99aa | ENSP00000469906 |
| HNRNPM-208 | ENST00000597270 | 1147 | 383aa | ENSP00000470635 |
| HNRNPM-207 | ENST00000597081 | 1004 | No protein | - |
| HNRNPM-211 | ENST00000598603 | 784 | No protein | - |
| HNRNPM-205 | ENST00000596295 | 910 | 214aa | ENSP00000472599 |
| HNRNPM-212 | ENST00000598999 | 409 | 116aa | ENSP00000469681 |
| HNRNPM-216 | ENST00000602219 | 1809 | No protein | - |
Phenotypes
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| 34007 | Tunica Media | 8.9730861E-005 | - |
| 52923 | Interleukin 1 Receptor Antagonist Protein | 5.8410000E-005 | - |
| 52931 | Interleukin 1 Receptor Antagonist Protein | 4.8500000E-005 | - |
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
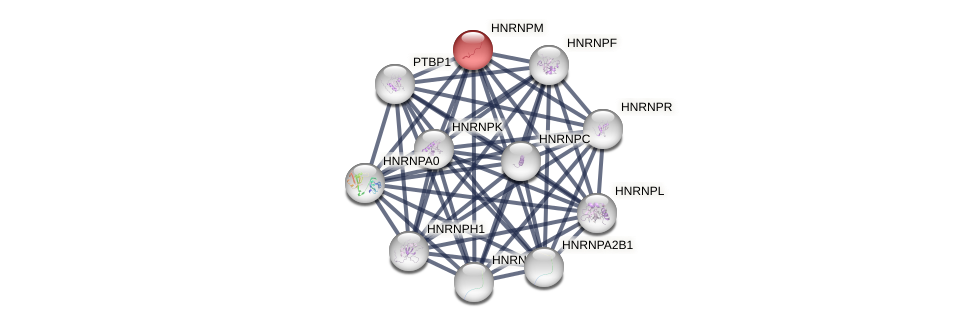
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000099783 | homo_sapiens | ENSP00000316950 | 67 | 53.5 | homo_sapiens | ENSP00000325376 | 55.0685 | 43.9726 |
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 13.0137 | 13.0137 | nomascus_leucogenys | ENSNLEP00000002256 | 95 | 95 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 99.589 | 99.589 | pan_paniscus | ENSPPAP00000033281 | 99.589 | 99.589 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 100 | 99.863 | pongo_abelii | ENSPPYP00000010664 | 100 | 99.863 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 99.589 | 99.589 | pan_troglodytes | ENSPTRP00000017736 | 99.589 | 99.589 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 99.589 | 99.589 | gorilla_gorilla | ENSGGOP00000018014 | 99.589 | 99.589 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 88.9041 | 83.2877 | ornithorhynchus_anatinus | ENSOANP00000009364 | 88.7825 | 83.1737 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 95.2055 | 90.6849 | sarcophilus_harrisii | ENSSHAP00000034485 | 93.5397 | 89.0983 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 91.7808 | 87.2603 | notamacropus_eugenii | ENSMEUP00000004376 | 92.2865 | 87.741 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 99.0411 | 98.4931 | choloepus_hoffmanni | ENSCHOP00000012063 | 99.0411 | 98.4931 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 99.0411 | 98.6301 | dasypus_novemcinctus | ENSDNOP00000009444 | 87.1084 | 86.747 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 47.2603 | 43.1507 | erinaceus_europaeus | ENSEEUP00000001770 | 63.4191 | 57.9044 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 96.0274 | 95.0685 | echinops_telfairi | ENSETEP00000016014 | 96.0274 | 95.0685 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 99.589 | 99.4521 | callithrix_jacchus | ENSCJAP00000020089 | 99.589 | 99.4521 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 99.589 | 99.589 | cercocebus_atys | ENSCATP00000024136 | 99.589 | 99.589 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 99.0411 | 99.0411 | macaca_fascicularis | ENSMFAP00000045533 | 99.4498 | 99.4498 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 100 | 100 | macaca_mulatta | ENSMMUP00000024753 | 100 | 100 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 100 | 100 | macaca_nemestrina | ENSMNEP00000026791 | 100 | 100 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 100 | 100 | papio_anubis | ENSPANP00000009850 | 100 | 100 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 99.589 | 99.589 | mandrillus_leucophaeus | ENSMLEP00000036242 | 99.589 | 99.589 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 94.9315 | 94.7945 | tursiops_truncatus | ENSTTRP00000009268 | 94.9315 | 94.7945 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 92.6027 | 91.9178 | vulpes_vulpes | ENSVVUP00000034990 | 89.6552 | 88.992 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 98.3562 | 97.6712 | mus_spretus | MGP_SPRETEiJ_P0045909 | 98.4911 | 97.8052 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 99.589 | 99.4521 | equus_asinus | ENSEASP00005032819 | 91.3317 | 91.206 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 93.9726 | 93.6986 | capra_hircus | ENSCHIP00000032304 | 99.2764 | 98.987 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 97.8082 | 96.7123 | loxodonta_africana | ENSLAFP00000011385 | 98.0769 | 96.978 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 99.4521 | 99.3151 | panthera_leo | ENSPLOP00000000932 | 99.4521 | 99.3151 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 99.4521 | 99.3151 | ailuropoda_melanoleuca | ENSAMEP00000005281 | 99.4521 | 99.3151 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 99.589 | 99.3151 | bos_indicus_hybrid | ENSBIXP00005030288 | 99.589 | 99.3151 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 85.6164 | 85.0685 | oryctolagus_cuniculus | ENSOCUP00000023086 | 98.4252 | 97.7953 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 99.589 | 99.4521 | equus_caballus | ENSECAP00000081627 | 91.3317 | 91.206 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 99.0411 | 98.9041 | canis_lupus_familiaris | ENSCAFP00845008810 | 92.4552 | 92.3274 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 99.0411 | 98.9041 | panthera_pardus | ENSPPRP00000017187 | 99.0411 | 98.9041 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 96.4384 | 96.3014 | marmota_marmota_marmota | ENSMMMP00000024693 | 96.4384 | 96.3014 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 99.589 | 99.4521 | balaenoptera_musculus | ENSBMSP00010018161 | 99.589 | 99.4521 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 99.3151 | 98.4931 | cavia_porcellus | ENSCPOP00000031827 | 99.3151 | 98.4931 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 99.0411 | 98.9041 | felis_catus | ENSFCAP00000014117 | 99.0411 | 98.9041 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 97.1233 | 96.7123 | ursus_americanus | ENSUAMP00000007688 | 91.8394 | 91.4508 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 43.4247 | 41.3699 | monodelphis_domestica | ENSMODP00000004247 | 87.0879 | 82.967 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 90.411 | 86.0274 | monodelphis_domestica | ENSMODP00000004654 | 98.2143 | 93.4524 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 94.2466 | 93.9726 | bison_bison_bison | ENSBBBP00000011661 | 94.2466 | 93.9726 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 90.137 | 88.4931 | monodon_monoceros | ENSMMNP00015027556 | 84.359 | 82.8205 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 93.9726 | 93.6986 | sus_scrofa | ENSSSCP00000014455 | 84.2752 | 84.0295 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 91.9178 | 91.3699 | microcebus_murinus | ENSMICP00000002312 | 89.1102 | 88.579 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 12.4658 | 12.1918 | octodon_degus | ENSODEP00000011424 | 86.6667 | 84.7619 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 83.8356 | 80.8219 | octodon_degus | ENSODEP00000012383 | 92.0301 | 88.7218 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 12.6027 | 12.1918 | octodon_degus | ENSODEP00000014514 | 93.8775 | 90.8163 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 13.8356 | 13.5616 | octodon_degus | ENSODEP00000023709 | 99.0196 | 97.0588 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 98.3562 | 97.3973 | jaculus_jaculus | ENSJJAP00000015087 | 98.3562 | 97.3973 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 99.589 | 99.3151 | ovis_aries_rambouillet | ENSOARP00020017252 | 99.589 | 99.3151 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 76.4384 | 75.8904 | ursus_maritimus | ENSUMAP00000019910 | 99.1119 | 98.4014 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 65.0685 | 63.9726 | ochotona_princeps | ENSOPRP00000001806 | 68.8406 | 67.6812 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 94.1096 | 93.9726 | delphinapterus_leucas | ENSDLEP00000019022 | 94.8895 | 94.7514 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 94.6575 | 94.5205 | physeter_catodon | ENSPCTP00005010954 | 99.8555 | 99.711 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 98.3562 | 97.8082 | mus_caroli | MGP_CAROLIEiJ_P0044005 | 98.4911 | 97.9424 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 98.3562 | 97.5342 | mus_musculus | ENSMUSP00000120115 | 98.4911 | 97.668 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 99.4521 | 99.0411 | dipodomys_ordii | ENSDORP00000007605 | 99.4521 | 99.0411 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 93.8356 | 93.6986 | mustela_putorius_furo | ENSMPUP00000007183 | 85.3051 | 85.1806 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 99.726 | 99.726 | aotus_nancymaae | ENSANAP00000015039 | 99.726 | 99.726 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 99.3151 | 99.1781 | saimiri_boliviensis_boliviensis | ENSSBOP00000032916 | 99.3151 | 99.1781 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 81.6438 | 81.3699 | vicugna_pacos | ENSVPAP00000003916 | 88.6905 | 88.3929 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 91.6438 | 90.8219 | chlorocebus_sabaeus | ENSCSAP00000004250 | 96.6763 | 95.8092 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 66.4384 | 66.1644 | tupaia_belangeri | ENSTBEP00000007557 | 70.9064 | 70.614 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 98.9041 | 98.3562 | mesocricetus_auratus | ENSMAUP00000019160 | 98.9041 | 98.3562 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 99.726 | 99.589 | phocoena_sinus | ENSPSNP00000018007 | 99.726 | 99.589 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 99.726 | 99.589 | camelus_dromedarius | ENSCDRP00005015346 | 99.726 | 99.589 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 83.9726 | 82.0548 | carlito_syrichta | ENSTSYP00000031573 | 93.4451 | 91.311 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 90.9589 | 90.8219 | panthera_tigris_altaica | ENSPTIP00000001275 | 94.3182 | 94.1761 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 13.6986 | 12.6027 | rattus_norvegicus | ENSRNOP00000069439 | 72.9927 | 67.1533 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 35.3425 | 33.8356 | prolemur_simus | ENSPSMP00000008223 | 94.1606 | 90.146 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 98.6301 | 98.0822 | microtus_ochrogaster | ENSMOCP00000006210 | 98.6301 | 98.0822 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 71.2329 | 70.6849 | sorex_araneus | ENSSARP00000000242 | 71.1354 | 70.5882 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 99.0411 | 98.4931 | peromyscus_maniculatus_bairdii | ENSPEMP00000007356 | 99.0411 | 98.4931 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 99.589 | 99.3151 | bos_mutus | ENSBMUP00000034613 | 99.589 | 99.3151 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 92.7397 | 92.0548 | mus_spicilegus | ENSMSIP00000009793 | 87.9221 | 87.2727 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 69.863 | 69.1781 | cricetulus_griseus_chok1gshd | ENSCGRP00001010047 | 92.3913 | 91.4855 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 12.8767 | 12.6027 | cricetulus_griseus_chok1gshd | ENSCGRP00001022820 | 94 | 92 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 89.0411 | 88.7671 | pteropus_vampyrus | ENSPVAP00000009094 | 98.6343 | 98.3308 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 93.8356 | 93.5616 | myotis_lucifugus | ENSMLUP00000006124 | 97.3011 | 97.017 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 72.0548 | 71.3699 | myotis_lucifugus | ENSMLUP00000017750 | 94.0966 | 93.2021 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 92.4658 | 88.0822 | vombatus_ursinus | ENSVURP00010032413 | 97.4026 | 92.785 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 68.3562 | 67.3973 | rhinolophus_ferrumequinum | ENSRFEP00010014807 | 97.4609 | 96.0938 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 99.4521 | 99.1781 | neovison_vison | ENSNVIP00000021962 | 99.4521 | 99.1781 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 99.589 | 99.3151 | bos_taurus | ENSBTAP00000074476 | 99.589 | 99.3151 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 13.9726 | 13.0137 | heterocephalus_glaber_female | ENSHGLP00000006365 | 77.8626 | 72.5191 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 90.5479 | 90 | rhinopithecus_bieti | ENSRBIP00000035755 | 99.5482 | 98.9458 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 95.0685 | 94.5205 | canis_lupus_dingo | ENSCAFP00020028289 | 88.6335 | 88.1226 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 30.137 | 30.137 | sciurus_vulgaris | ENSSVLP00005008239 | 100 | 100 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 99.589 | 99.4521 | sciurus_vulgaris | ENSSVLP00005024085 | 99.589 | 99.4521 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 14.1096 | 13.5616 | sciurus_vulgaris | ENSSVLP00005008223 | 82.4 | 79.2 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 90.411 | 86.0274 | phascolarctos_cinereus | ENSPCIP00000027132 | 98.2143 | 93.4524 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 98.6301 | 97.2603 | procavia_capensis | ENSPCAP00000009046 | 98.6301 | 97.2603 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 69.1781 | 64.9315 | nannospalax_galili | ENSNGAP00000017150 | 89.698 | 84.1918 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 10.5479 | 9.58904 | nannospalax_galili | ENSNGAP00000004176 | 88.5057 | 80.4598 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 92.6027 | 91.2329 | bos_grunniens | ENSBGRP00000040208 | 91.1051 | 89.7574 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 75.2055 | 70.9589 | chinchilla_lanigera | ENSCLAP00000004287 | 88.5484 | 83.5484 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 99.589 | 99.3151 | cervus_hanglu_yarkandensis | ENSCHYP00000017631 | 99.589 | 99.3151 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 99.589 | 99.3151 | catagonus_wagneri | ENSCWAP00000022327 | 99.589 | 99.3151 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 99.589 | 99.4521 | ictidomys_tridecemlineatus | ENSSTOP00000029033 | 99.589 | 99.4521 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 40.137 | 37.1233 | otolemur_garnettii | ENSOGAP00000021070 | 88.5196 | 81.8731 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 99.0411 | 99.0411 | cebus_imitator | ENSCCAP00000004462 | 99.5868 | 99.5868 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 96.9863 | 96.8493 | urocitellus_parryii | ENSUPAP00010005847 | 90.653 | 90.525 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 88.3562 | 88.0822 | moschus_moschiferus | ENSMMSP00000006031 | 98.9264 | 98.6196 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 99.589 | 99.3151 | moschus_moschiferus | ENSMMSP00000004741 | 99.589 | 99.3151 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 99.3151 | 99.1781 | rhinopithecus_roxellana | ENSRROP00000029024 | 99.3151 | 99.1781 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 92.0548 | 91.9178 | rhinopithecus_roxellana | ENSRROP00000018242 | 98.2456 | 98.0994 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 98.2192 | 97.6712 | mus_pahari | MGP_PahariEiJ_P0043562 | 98.3539 | 97.8052 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 93.5616 | 93.1507 | propithecus_coquereli | ENSPCOP00000016976 | 90.5836 | 90.1857 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 29.589 | 21.3699 | caenorhabditis_elegans | C25A1.4.1 | 47.5771 | 34.3612 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 36.7123 | 25.4795 | drosophila_melanogaster | FBpp0081601 | 42.4051 | 29.4304 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 36.5753 | 24.6575 | ciona_intestinalis | ENSCINP00000028897 | 52.4558 | 35.3635 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 55.2055 | 43.9726 | callorhinchus_milii | ENSCMIP00000021095 | 68.0743 | 54.223 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 61.5069 | 48.0822 | latimeria_chalumnae | ENSLACP00000019111 | 72.4194 | 56.6129 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 68.3562 | 55.7534 | lepisosteus_oculatus | ENSLOCP00000005330 | 76.2997 | 62.2324 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 61.9178 | 51.7808 | clupea_harengus | ENSCHAP00000043344 | 69.5385 | 58.1538 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 66.3014 | 54.2466 | danio_rerio | ENSDARP00000121443 | 67.6923 | 55.3846 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 66.8493 | 54.9315 | carassius_auratus | ENSCARP00000024746 | 72.619 | 59.6726 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 66.7123 | 54.7945 | carassius_auratus | ENSCARP00000084714 | 70.2742 | 57.7201 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 66.8493 | 54.9315 | carassius_auratus | ENSCARP00000018407 | 72.619 | 59.6726 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 64.9315 | 53.8356 | astyanax_mexicanus | ENSAMXP00000016570 | 70.1183 | 58.1361 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 65.0685 | 53.8356 | ictalurus_punctatus | ENSIPUP00000037516 | 72.8528 | 60.2761 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 64.1096 | 53.6986 | esox_lucius | ENSELUP00000040503 | 69.5394 | 58.2467 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 63.0137 | 52.0548 | electrophorus_electricus | ENSEEEP00000035104 | 69.8027 | 57.6631 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 62.7397 | 52.6027 | oncorhynchus_kisutch | ENSOKIP00005014993 | 68.8722 | 57.7444 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 63.8356 | 53.5616 | oncorhynchus_kisutch | ENSOKIP00005007722 | 71.4724 | 59.9693 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 64.3836 | 53.5616 | oncorhynchus_mykiss | ENSOMYP00000117730 | 67.7233 | 56.3401 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 63.8356 | 52.7397 | oncorhynchus_mykiss | ENSOMYP00000036188 | 69.8651 | 57.7211 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 65.6164 | 55.0685 | salmo_salar | ENSSSAP00000062775 | 71.9219 | 60.3604 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 64.7945 | 53.8356 | salmo_salar | ENSSSAP00000167292 | 69.764 | 57.9646 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 62.7397 | 51.7808 | salmo_trutta | ENSSTUP00000023405 | 70.4615 | 58.1538 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 63.2877 | 53.1507 | salmo_trutta | ENSSTUP00000093252 | 69.3694 | 58.2583 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 59.1781 | 47.9452 | gadus_morhua | ENSGMOP00000010398 | 67.5 | 54.6875 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 65.3425 | 54.5205 | fundulus_heteroclitus | ENSFHEP00000005726 | 68.2403 | 56.9385 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 65.7534 | 54.2466 | poecilia_reticulata | ENSPREP00000023868 | 66.39 | 54.7718 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 58.3562 | 47.6712 | xiphophorus_maculatus | ENSXMAP00000037770 | 67.4051 | 55.0633 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 62.7397 | 52.1918 | oryzias_latipes | ENSORLP00000028445 | 68.5629 | 57.0359 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 63.6986 | 52.3288 | cyclopterus_lumpus | ENSCLMP00005049231 | 67.1965 | 55.2023 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 65.6164 | 55.0685 | oreochromis_niloticus | ENSONIP00000002202 | 70.5449 | 59.2047 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 63.9726 | 53.6986 | haplochromis_burtoni | ENSHBUP00000035324 | 72.0679 | 60.4938 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 62.8767 | 52.4658 | astatotilapia_calliptera | ENSACLP00000030503 | 69.4402 | 57.9425 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 66.1644 | 54.5205 | sparus_aurata | ENSSAUP00010067674 | 68.8034 | 56.6952 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 62.3288 | 50.137 | lates_calcarifer | ENSLCAP00010050048 | 64.0845 | 51.5493 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 74.2466 | 60 | xenopus_tropicalis | ENSXETP00000003692 | 72.8495 | 58.871 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 9.45205 | 8.35616 | chrysemys_picta_bellii | ENSCPBP00000023921 | 83.1325 | 73.494 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 75.0685 | 67.1233 | chrysemys_picta_bellii | ENSCPBP00000023919 | 88.6731 | 79.288 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 80.5479 | 71.2329 | sphenodon_punctatus | ENSSPUP00000002943 | 86.3436 | 76.3583 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 76.3014 | 66.0274 | laticauda_laticaudata | ENSLLTP00000000655 | 84.139 | 72.8097 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 43.4247 | 35.0685 | notechis_scutatus | ENSNSUP00000001299 | 79.4486 | 64.1604 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 43.4247 | 35.0685 | notechis_scutatus | ENSNSUP00000001296 | 79.4486 | 64.1604 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 25.2055 | 20 | notechis_scutatus | ENSNSUP00000001286 | 88.4615 | 70.1923 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 25.2055 | 20 | notechis_scutatus | ENSNSUP00000001291 | 88.4615 | 70.1923 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 74.5205 | 63.9726 | pseudonaja_textilis | ENSPTXP00000023743 | 83.4356 | 71.6258 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 79.589 | 70.5479 | anas_platyrhynchos_platyrhynchos | ENSAPLP00000009883 | 84.8175 | 75.1825 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 88.3562 | 79.4521 | gallus_gallus | ENSGALP00010042185 | 83.5492 | 75.1295 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 85.3425 | 76.8493 | meleagris_gallopavo | ENSMGAP00000000090 | 93.4033 | 84.1079 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 74.5205 | 66.5753 | serinus_canaria | ENSSCAP00000017284 | 79.1849 | 70.7424 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 80.6849 | 71.9178 | parus_major | ENSPMJP00000017059 | 83.6648 | 74.5739 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 66.1644 | 55.3425 | neolamprologus_brichardi | ENSNBRP00000020916 | 70.8211 | 59.2375 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 84.3836 | 73.4247 | naja_naja | ENSNNAP00000004271 | 82.9071 | 72.14 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 76.1644 | 65.4795 | podarcis_muralis | ENSPMRP00000034664 | 86.6044 | 74.4548 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 12.8767 | 10.9589 | podarcis_muralis | ENSPMRP00000034650 | 58.0247 | 49.3827 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 66.5753 | 60.8219 | geospiza_fortis | ENSGFOP00000013235 | 86.631 | 79.1444 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 85.8904 | 77.2603 | taeniopygia_guttata | ENSTGUP00000000966 | 72.3183 | 65.0519 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 65.0685 | 53.9726 | erpetoichthys_calabaricus | ENSECRP00000030671 | 73.9875 | 61.3707 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 65.7534 | 54.2466 | kryptolebias_marmoratus | ENSKMAP00000029947 | 69.5652 | 57.3913 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 71.2329 | 61.0959 | salvator_merianae | ENSSMRP00000025223 | 85.8086 | 73.5974 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 12.1918 | 11.0959 | salvator_merianae | ENSSMRP00000025224 | 62.2378 | 56.6434 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 62.8767 | 52.8767 | oryzias_javanicus | ENSOJAP00000042909 | 71.2733 | 59.9379 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 66.4384 | 55.7534 | amphilophus_citrinellus | ENSACIP00000015424 | 71.1144 | 59.6774 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 66.7123 | 54.9315 | dicentrarchus_labrax | ENSDLAP00005015305 | 69.1761 | 56.9602 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 66.8493 | 55.0685 | amphiprion_percula | ENSAPEP00000005922 | 71.5543 | 58.9443 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 65.7534 | 54.5205 | oryzias_sinensis | ENSOSIP00000010950 | 69.7674 | 57.8488 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 79.863 | 72.4658 | pelodiscus_sinensis | ENSPSIP00000012893 | 87.4063 | 79.3103 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 10.274 | 9.45205 | gopherus_evgoodei | ENSGEVP00005023872 | 79.7872 | 73.4043 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 77.8082 | 68.9041 | gopherus_evgoodei | ENSGEVP00005023906 | 86.7176 | 76.7939 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 56.8493 | 49.4521 | chelonoidis_abingdonii | ENSCABP00000006081 | 89.6328 | 77.9698 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 10.411 | 9.45205 | chelonoidis_abingdonii | ENSCABP00000004560 | 76.7677 | 69.697 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 51.5069 | 43.0137 | seriola_dumerili | ENSSDUP00000002880 | 74.1617 | 61.9329 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 63.4247 | 52.1918 | cottoperca_gobio | ENSCGOP00000027309 | 69.6241 | 57.2932 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 85.7534 | 77.2603 | struthio_camelus_australis | ENSSCUP00000003050 | 93.7126 | 84.4311 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 81.3699 | 72.3288 | anser_brachyrhynchus | ENSABRP00000014685 | 84.136 | 74.7875 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 61.5069 | 50.6849 | gasterosteus_aculeatus | ENSGACP00000013576 | 68.6544 | 56.5749 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 64.5205 | 52.7397 | cynoglossus_semilaevis | ENSCSEP00000023491 | 68.2609 | 55.7971 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 55.7534 | 45.6164 | poecilia_formosa | ENSPFOP00000013730 | 70.1724 | 57.4138 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 67.3973 | 56.4384 | myripristis_murdjan | ENSMMDP00005023358 | 70.3863 | 58.9413 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 66.5753 | 55.0685 | sander_lucioperca | ENSSLUP00000043712 | 70.0288 | 57.9251 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 80.9589 | 72.4658 | strigops_habroptila | ENSSHBP00005020070 | 87.426 | 78.2544 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 65.7534 | 54.3836 | amphiprion_ocellaris | ENSAOCP00000005063 | 72.9483 | 60.3343 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 85.4795 | 77.2603 | terrapene_carolina_triunguis | ENSTMTP00000026027 | 93.5532 | 84.5577 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 65.3425 | 54.7945 | hucho_hucho | ENSHHUP00000047018 | 71.5142 | 59.97 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 64.6575 | 53.9726 | hucho_hucho | ENSHHUP00000024992 | 70.1337 | 58.5438 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 64.3836 | 53.2877 | betta_splendens | ENSBSLP00000033648 | 70.5706 | 58.4084 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 64.2466 | 53.9726 | pygocentrus_nattereri | ENSPNAP00000032544 | 71.7125 | 60.2446 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 65.6164 | 54.2466 | anabas_testudineus | ENSATEP00000013001 | 71.8141 | 59.3703 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 36.4384 | 24.6575 | ciona_savignyi | ENSCSAVP00000001355 | 55.7652 | 37.7358 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 22.7397 | 14.1096 | saccharomyces_cerevisiae | YCL011C | 38.8759 | 24.1218 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 20.274 | 11.6438 | saccharomyces_cerevisiae | YNL004W | 32.5991 | 18.7225 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 63.0137 | 52.4658 | seriola_lalandi_dorsalis | ENSSLDP00000021544 | 67.9468 | 56.5731 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 65.7534 | 53.6986 | labrus_bergylta | ENSLBEP00000005894 | 67.3212 | 54.979 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 62.7397 | 52.3288 | pundamilia_nyererei | ENSPNYP00000008768 | 68.8722 | 57.4436 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 65.0685 | 53.8356 | mastacembelus_armatus | ENSMAMP00000008241 | 70.3704 | 58.2222 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 64.3836 | 53.2877 | stegastes_partitus | ENSSPAP00000030869 | 71.8654 | 59.4801 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 63.9726 | 53.6986 | oncorhynchus_tshawytscha | ENSOTSP00005071189 | 71.6258 | 60.1227 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 64.1096 | 53.5616 | oncorhynchus_tshawytscha | ENSOTSP00005073704 | 70.0599 | 58.5329 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 60 | 48.9041 | acanthochromis_polyacanthus | ENSAPOP00000021093 | 69.5238 | 56.6667 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 64.5205 | 52.3288 | nothobranchius_furzeri | ENSNFUP00015054871 | 62.2193 | 50.4624 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 65.8904 | 53.8356 | cyprinodon_variegatus | ENSCVAP00000017708 | 69.8113 | 57.0392 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 63.9726 | 53.2877 | denticeps_clupeoides | ENSDCDP00000051722 | 70.9726 | 59.1185 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 66.1644 | 54.1096 | larimichthys_crocea | ENSLCRP00005057632 | 68.608 | 56.108 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 65.3425 | 52.8767 | takifugu_rubripes | ENSTRUP00000069160 | 69.1304 | 55.942 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 80.274 | 71.3699 | ficedula_albicollis | ENSFALP00000001105 | 85.1744 | 75.7267 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 63.8356 | 52.3288 | hippocampus_comes | ENSHCOP00000016203 | 72.3602 | 59.3168 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 67.5342 | 55.6164 | cyprinus_carpio_carpio | ENSCCRP00000108934 | 73.4724 | 60.5067 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 67.1233 | 55.6164 | cyprinus_carpio_carpio | ENSCCRP00000135164 | 73.3533 | 60.7784 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 82.8767 | 74.1096 | coturnix_japonica | ENSCJPP00005006472 | 85.8156 | 76.7376 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 63.5616 | 52.1918 | poecilia_latipinna | ENSPLAP00000008475 | 69.5652 | 57.1214 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 65.6164 | 53.1507 | sinocyclocheilus_grahami | ENSSGRP00000056108 | 70.0292 | 56.7251 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 63.4247 | 51.6438 | sinocyclocheilus_grahami | ENSSGRP00000108454 | 67.4927 | 54.9563 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 66.9863 | 56.1644 | maylandia_zebra | ENSMZEP00005003236 | 70.1578 | 58.8235 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 64.2466 | 51.7808 | tetraodon_nigroviridis | ENSTNIP00000018601 | 70 | 56.4179 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 65.4795 | 54.6575 | paramormyrops_kingsleyae | ENSPKIP00000014164 | 73.8794 | 61.6692 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 65.2055 | 54.1096 | scleropages_formosus | ENSSFOP00015013912 | 73.3436 | 60.8629 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 57.1233 | 43.8356 | leptobrachium_leishanense | ENSLLEP00000007275 | 62.3318 | 47.8326 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 66.7123 | 55.2055 | scophthalmus_maximus | ENSSMAP00000006457 | 68.4951 | 56.6807 |
| ENSG00000099783 | homo_sapiens | ENSP00000325376 | 66.1644 | 55.0685 | oryzias_melastigma | ENSOMEP00000011752 | 69.1977 | 57.5931 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0000380 | involved_in alternative mRNA splicing, via spliceosome | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0000398 | involved_in mRNA splicing, via spliceosome | 1 | IC | Homo_sapiens(9606) | Process |
| GO:0003723 | enables RNA binding | 2 | HDA,NAS | Homo_sapiens(9606) | Function |
| GO:0003729 | enables mRNA binding | 1 | IBA | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005634 | is_active_in nucleus | 1 | IBA | Homo_sapiens(9606) | Component |
| GO:0005654 | located_in nucleoplasm | 2 | IDA,TAS | Homo_sapiens(9606) | Component |
| GO:0005681 | part_of spliceosomal complex | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005730 | located_in nucleolus | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0016020 | located_in membrane | 1 | HDA | Homo_sapiens(9606) | Component |
| GO:0016363 | located_in nuclear matrix | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0019904 | enables protein domain specific binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0042382 | located_in paraspeckles | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0045202 | located_in synapse | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0062023 | located_in collagen-containing extracellular matrix | 1 | HDA | Homo_sapiens(9606) | Component |
| GO:0070062 | located_in extracellular exosome | 1 | HDA | Homo_sapiens(9606) | Component |
| GO:0071013 | part_of catalytic step 2 spliceosome | 1 | IDA | Homo_sapiens(9606) | Component |