RBP Type
Non-canonical_RBPs
Diseases
CHIKV SARS-COV-OC43 Flaviviruses HIV IAV SARS-COV-2 VEEV Dengue Zika
Drug
N.A.
Main interacting RNAs
N.A.
Moonlighting functions
N.A.
Localizations
Stress granucle
BulkPerturb-seq
N.A.
Ensembl ID ENSG00000047849 Gene ID
4134 Accession
6862
Symbol
MAP4
Alias
NA
Full Name
microtubule associated protein 4
Status
Confidence
Length
238583 bases
Strand
Minus strand
Position
3 : 47850690 - 48089272
RNA binding domain
N.A.
Summary
The protein encoded by this gene is a major non-neuronal microtubule-associated protein. This protein contains a domain similar to the microtubule-binding domains of neuronal microtubule-associated protein (MAP2) and microtubule-associated protein tau (MAPT/TAU). This protein promotes microtubule assembly, and has been shown to counteract destabilization of interphase microtubule catastrophe promotion. Cyclin B was found to interact with this protein, which targets cell division cycle 2 (CDC2) kinase to microtubules. The phosphorylation of this protein affects microtubule properties and cell cycle progression. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Aug 2008]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
Name
Transcript ID
bp
Protein
Translation ID
MAP4-208
ENST00000429422
3724
493aa
ENSP00000416743
MAP4-217
ENST00000683076
9232
2296aa
ENSP00000507895
MAP4-204
ENST00000395734
5600
1135aa
ENSP00000379083
MAP4-207
ENST00000426837
8920
2297aa
ENSP00000407602
MAP4-202
ENST00000360240
5142
1152aa
ENSP00000353375
MAP4-205
ENST00000420772
3478
828aa
ENSP00000409731
MAP4-201
ENST00000335271
1564
463aa
ENSP00000334770
MAP4-213
ENST00000477765
284
No protein
-
MAP4-203
ENST00000383736
2282
760aa
ENSP00000373242
MAP4-211
ENST00000462206
572
No protein
-
MAP4-215
ENST00000497735
2156
No protein
-
MAP4-214
ENST00000482752
386
No protein
-
MAP4-206
ENST00000423088
899
283aa
ENSP00000415689
MAP4-216
ENST00000633710
570
140aa
ENSP00000487797
MAP4-209
ENST00000434267
2772
99aa
ENSP00000402767
MAP4-210
ENST00000439356
2743
99aa
ENSP00000397414
MAP4-212
ENST00000468075
487
76aa
ENSP00000487698
Pathway ID
Pathway Name
Source
ensgID
Trait
pValue
Pubmed ID
17777 Alcoholism 7.45338e-005 22978509 17820 Alcoholism 7.39231e-005 22978509 62300 Arterial Pressure 3E-8 21909110 88239 Education 8E-9 27225129
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
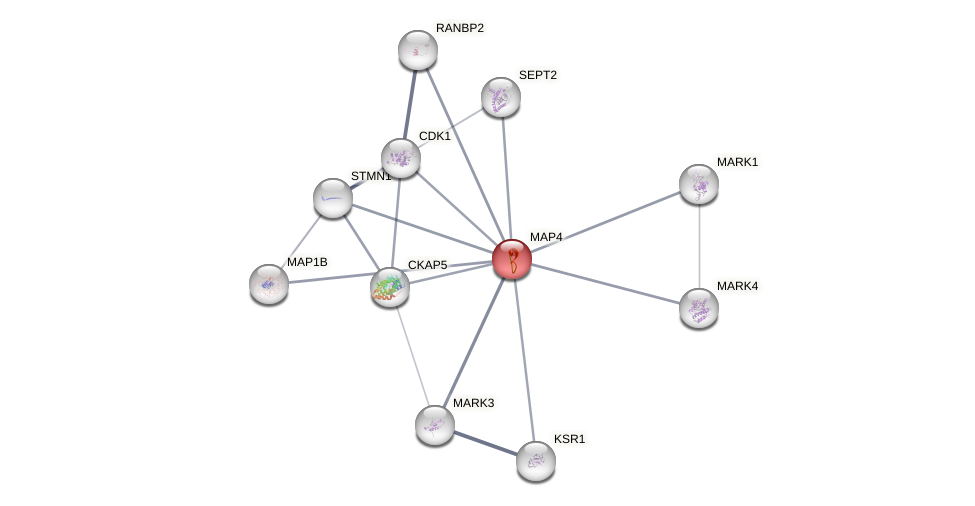
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
ENSG00000047849
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
ENSG00000047849 homo_sapiens ENSP00000507895 98.6063 98.2143 pan_paniscus ENSPPAP00000039205 98.5633 98.1715 ENSG00000047849 homo_sapiens ENSP00000507895 95.993 94.9042 pongo_abelii ENSPPYP00000044232 97.0498 95.9489 ENSG00000047849 homo_sapiens ENSP00000507895 98.7369 98.2578 pan_troglodytes ENSPTRP00000073554 98.4796 98.0017 ENSG00000047849 homo_sapiens ENSP00000507895 98.3885 97.7787 gorilla_gorilla ENSGGOP00000040115 98.2601 97.6512 ENSG00000047849 homo_sapiens ENSP00000507895 77.047 67.2038 dasypus_novemcinctus ENSDNOP00000018562 79.3986 69.2549 ENSG00000047849 homo_sapiens ENSP00000507895 83.3624 79.7038 callithrix_jacchus ENSCJAP00000089154 89.6487 85.7143 ENSG00000047849 homo_sapiens ENSP00000507895 95.1655 93.2491 cercocebus_atys ENSCATP00000018233 96.8958 94.9446 ENSG00000047849 homo_sapiens ENSP00000507895 95.2962 93.5976 macaca_fascicularis ENSMFAP00000015943 97.0719 95.3416 ENSG00000047849 homo_sapiens ENSP00000507895 96.2108 94.4686 macaca_mulatta ENSMMUP00000039163 96.9285 95.1733 ENSG00000047849 homo_sapiens ENSP00000507895 95.4268 93.5105 macaca_nemestrina ENSMNEP00000004365 97.1619 95.2106 ENSG00000047849 homo_sapiens ENSP00000507895 94.0767 91.8118 papio_anubis ENSPANP00000047835 94.3231 92.0524 ENSG00000047849 homo_sapiens ENSP00000507895 95.4268 93.5105 mandrillus_leucophaeus ENSMLEP00000004883 97.1619 95.2106 ENSG00000047849 homo_sapiens ENSP00000507895 43.8153 36.3676 mus_spretus MGP_SPRETEiJ_P0093922 69.5231 57.7056 ENSG00000047849 homo_sapiens ENSP00000507895 36.1498 31.1411 capra_hircus ENSCHIP00000014056 74.2397 63.9535 ENSG00000047849 homo_sapiens ENSP00000507895 35.9321 30.7491 loxodonta_africana ENSLAFP00000019013 75.2737 64.4161 ENSG00000047849 homo_sapiens ENSP00000507895 78.2665 69.9477 ailuropoda_melanoleuca ENSAMEP00000027035 74.2869 66.3911 ENSG00000047849 homo_sapiens ENSP00000507895 76.6551 67.6829 bos_indicus_hybrid ENSBIXP00005038203 79.6741 70.3486 ENSG00000047849 homo_sapiens ENSP00000507895 76.3502 66.7247 oryctolagus_cuniculus ENSOCUP00000038358 73.7795 64.4781 ENSG00000047849 homo_sapiens ENSP00000507895 80.5749 72.9965 equus_caballus ENSECAP00000041559 83.2958 75.4615 ENSG00000047849 homo_sapiens ENSP00000507895 76.2195 67.9878 canis_lupus_familiaris ENSCAFP00845010544 77.9163 69.5013 ENSG00000047849 homo_sapiens ENSP00000507895 75.6533 66.8554 sus_scrofa ENSSSCP00000036131 78.2432 69.1441 ENSG00000047849 homo_sapiens ENSP00000507895 43.8589 36.4111 mus_caroli MGP_CAROLIEiJ_P0090195 69.4962 57.695 ENSG00000047849 homo_sapiens ENSP00000507895 43.5976 36.1063 mus_musculus ENSMUSP00000143171 69.4657 57.5295 ENSG00000047849 homo_sapiens ENSP00000507895 89.8519 86.1063 aotus_nancymaae ENSANAP00000015409 91.364 87.5554 ENSG00000047849 homo_sapiens ENSP00000507895 90.0261 86.1063 saimiri_boliviensis_boliviensis ENSSBOP00000034823 91.4602 87.4779 ENSG00000047849 homo_sapiens ENSP00000507895 95.3833 93.5976 chlorocebus_sabaeus ENSCSAP00000012033 96.7742 94.9624 ENSG00000047849 homo_sapiens ENSP00000507895 76.4373 68.5105 camelus_dromedarius ENSCDRP00005014384 79.5197 71.2732 ENSG00000047849 homo_sapiens ENSP00000507895 76.5679 67.6829 bos_mutus ENSBMUP00000014949 79.5835 70.3486 ENSG00000047849 homo_sapiens ENSP00000507895 31.6638 26.1324 myotis_lucifugus ENSMLUP00000021442 67.2525 55.5042 ENSG00000047849 homo_sapiens ENSP00000507895 71.385 62.4129 rhinolophus_ferrumequinum ENSRFEP00010014954 70.0427 61.2393 ENSG00000047849 homo_sapiens ENSP00000507895 76.5679 67.5523 bos_taurus ENSBTAP00000056739 79.4038 70.0542 ENSG00000047849 homo_sapiens ENSP00000507895 73.5192 63.3275 heterocephalus_glaber_female ENSHGLP00000039276 75.6951 65.2018 ENSG00000047849 homo_sapiens ENSP00000507895 76.5679 67.6829 bos_grunniens ENSBGRP00000029854 79.5835 70.3486 ENSG00000047849 homo_sapiens ENSP00000507895 75.9582 67.8136 cervus_hanglu_yarkandensis ENSCHYP00000038487 79.562 71.031 ENSG00000047849 homo_sapiens ENSP00000507895 90.4181 86.8031 cebus_imitator ENSCCAP00000016659 91.8991 88.2249 ENSG00000047849 homo_sapiens ENSP00000507895 94.7735 93.0749 rhinopithecus_roxellana ENSRROP00000027698 95.9013 94.1825
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0000226 involved_in microtubule cytoskeleton organization 3 IBA,IDA,ISS Homo_sapiens(9606) Process GO:0003723 enables RNA binding 1 HDA Homo_sapiens(9606) Function GO:0005198 enables structural molecule activity 1 TAS Homo_sapiens(9606) Function GO:0005515 enables protein binding 1 IPI Homo_sapiens(9606) Function GO:0005815 located_in microtubule organizing center 1 IDA Homo_sapiens(9606) Component GO:0005874 is_active_in microtubule 1 IDA Homo_sapiens(9606) Component GO:0005875 part_of microtubule associated complex 1 TAS Homo_sapiens(9606) Component GO:0005930 located_in axoneme 1 IDA Homo_sapiens(9606) Component GO:0007052 acts_upstream_of_or_within mitotic spindle organization 1 IMP Homo_sapiens(9606) Process GO:0008017 enables microtubule binding 1 IBA Homo_sapiens(9606) Function GO:0030424 located_in axon 1 ISS Homo_sapiens(9606) Component GO:0031175 involved_in neuron projection development 1 IBA Homo_sapiens(9606) Process GO:0043005 is_active_in neuron projection 1 IBA Homo_sapiens(9606) Component GO:0046785 involved_in microtubule polymerization 1 IDA Homo_sapiens(9606) Process GO:0051012 acts_upstream_of_or_within microtubule sliding 1 IMP Homo_sapiens(9606) Process GO:0051294 acts_upstream_of_or_within establishment of spindle orientation 2 IGI,IMP Homo_sapiens(9606) Process GO:0051301 acts_upstream_of_or_within cell division 1 IMP Homo_sapiens(9606) Process GO:0061523 involved_in cilium disassembly 1 IDA Homo_sapiens(9606) Process GO:0072686 located_in mitotic spindle 1 IDA Homo_sapiens(9606) Component GO:0140778 enables microtubule stabilizing activity 1 IDA Homo_sapiens(9606) Function GO:1902856 involved_in negative regulation of non-motile cilium assembly 1 IMP Homo_sapiens(9606) Process
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.