Welcome to
RBP World!
Description
| Ensembl ID | ENSG00000214274 | Gene ID | 283 | Accession | 483 |
| Symbol | ANG | Alias | ALS9;RAA1;HEL168;RNASE4;RNASE5 | Full Name | angiogenin |
| Status | Confidence | Length | 10010 bases | Strand | Plus strand |
| Position | 14 : 20684177 - 20694186 | RNA binding domain | RnaseA | ||
| Summary | The protein encoded by this gene is a member of the RNase A superfamily though it has relatively weak ribonucleolytic activity. This protein is a potent mediator of new blood vessel formation and thus, in addition to the name RNase5, is commonly called angiogenin. This protein induces angiogenesis after binding to actin on the surface of endothelial cells. This protein also accumulates at the nucleolus where it stimulates ribosomal transcription. Under stress conditions this protein translocates to the cytosol where it hydrolyzes cellular tRNAs and influences protein synthesis. A signal peptide is cleaved from the precursor protein to produce a mature protein which contains a nuclear localization signal, a cell binding motif, and a catalytic domain. This protein has been shown to be both neurotrophic and neuroprotective and the mature protein has antimicrobial activity against some bacteria and fungi, including S. pneumoniae and C. albicans. Due to its effect on rRNA production and angiogenesis this gene plays important roles in cell growth and tumor progression. Mutations in this gene are associated with progression of amyotrophic lateral sclerosis (ALS). This gene and the neighboring RNase4 gene share promoters and 5' exons though each gene then splices to a distinct 3' exon containing the complete coding region of each gene. Alternative splicing results in multiple transcript variants encoding the same protein. [provided by RefSeq, Jul 2020] | ||||
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| ANG-201 | ENST00000336811 | 1222 | 147aa | ENSP00000336762 |
| ANG-203 | ENST00000554073 | 251 | No protein | - |
| ANG-202 | ENST00000397990 | 748 | 147aa | ENSP00000381077 |
Phenotypes
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| 15809 | Stroke | 2.1727993E-004 | - |
| 54574 | Cryptoxanthins | 8.3140000E-005 | - |
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
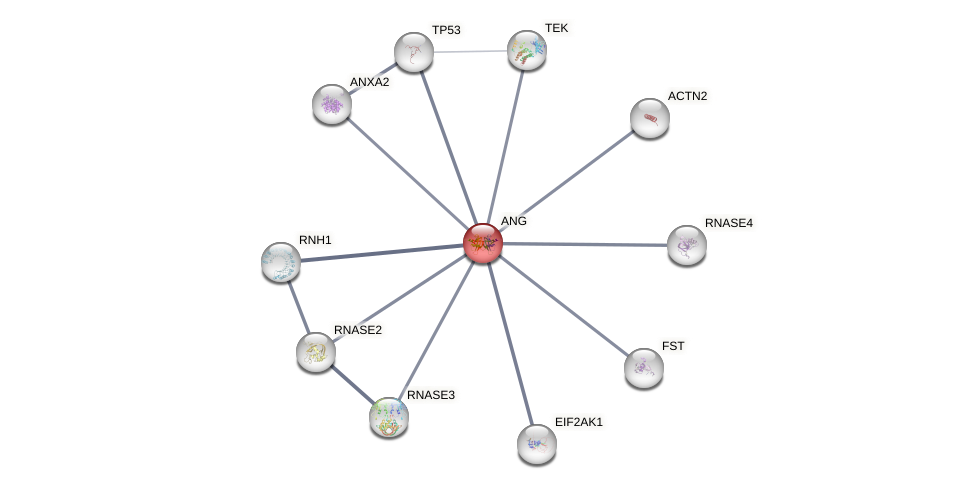
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000214274 | homo_sapiens | ENSP00000512867 | 31.2925 | 20.4082 | homo_sapiens | ENSP00000381077 | 31.2925 | 20.4082 |
| ENSG00000214274 | homo_sapiens | ENSP00000298690 | 39.7436 | 28.8462 | homo_sapiens | ENSP00000381077 | 42.1769 | 30.6122 |
| ENSG00000214274 | homo_sapiens | ENSP00000311398 | 35.7143 | 25.3247 | homo_sapiens | ENSP00000381077 | 37.415 | 26.5306 |
| ENSG00000214274 | homo_sapiens | ENSP00000302046 | 37.3333 | 26 | homo_sapiens | ENSP00000381077 | 38.0952 | 26.5306 |
| ENSG00000214274 | homo_sapiens | ENSP00000392996 | 24.0741 | 13.8889 | homo_sapiens | ENSP00000381077 | 35.3741 | 20.4082 |
| ENSG00000214274 | homo_sapiens | ENSP00000451318 | 22.1106 | 11.5578 | homo_sapiens | ENSP00000381077 | 29.932 | 15.6463 |
| ENSG00000214274 | homo_sapiens | ENSP00000384683 | 21.9048 | 10.9524 | homo_sapiens | ENSP00000381077 | 31.2925 | 15.6463 |
| ENSG00000214274 | homo_sapiens | ENSP00000381057 | 44.2308 | 31.4103 | homo_sapiens | ENSP00000381077 | 46.9388 | 33.3333 |
| ENSG00000214274 | homo_sapiens | ENSP00000452245 | 47.619 | 37.415 | homo_sapiens | ENSP00000381077 | 47.619 | 37.415 |
| ENSG00000214274 | homo_sapiens | ENSP00000372410 | 33.3333 | 19.2308 | homo_sapiens | ENSP00000381077 | 35.3741 | 20.4082 |
| ENSG00000214274 | homo_sapiens | ENSP00000302324 | 31.25 | 21.875 | homo_sapiens | ENSP00000381077 | 34.0136 | 23.8095 |
| ENSG00000214274 | homo_sapiens | ENSP00000303276 | 34.1615 | 22.3602 | homo_sapiens | ENSP00000381077 | 37.415 | 24.4898 |
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 98.6395 | 98.6395 | pan_paniscus | ENSPPAP00000019258 | 98.6395 | 98.6395 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 99.3197 | 99.3197 | pan_troglodytes | ENSPTRP00000066875 | 99.3197 | 99.3197 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 100 | 99.3197 | gorilla_gorilla | ENSGGOP00000035667 | 100 | 99.3197 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 77.551 | 61.2245 | sarcophilus_harrisii | ENSSHAP00000024006 | 76 | 60 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 71.4286 | 58.5034 | notamacropus_eugenii | ENSMEUP00000000624 | 73.9437 | 60.5634 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 83.6735 | 75.5102 | macaca_fascicularis | ENSMFAP00000051996 | 84.2466 | 76.0274 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 84.3537 | 76.1905 | macaca_fascicularis | ENSMFAP00000063079 | 58.7678 | 53.0806 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 82.3129 | 74.1497 | macaca_mulatta | ENSMMUP00000062606 | 82.8767 | 74.6575 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 76.8708 | 63.9456 | tursiops_truncatus | ENSTTRP00000008995 | 77.931 | 64.8276 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 74.1497 | 63.9456 | mus_spretus | MGP_SPRETEiJ_P0043290 | 75.1724 | 64.8276 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 74.1497 | 63.9456 | mus_spretus | MGP_SPRETEiJ_P0043289 | 75.1724 | 64.8276 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 83.6735 | 72.1088 | mus_spretus | MGP_SPRETEiJ_P0038221 | 84.8276 | 73.1034 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 85.034 | 74.8299 | equus_asinus | ENSEASP00005061523 | 85.6164 | 75.3425 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 77.551 | 64.6258 | capra_hircus | ENSCHIP00000006472 | 77.027 | 64.1892 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 74.8299 | 62.585 | capra_hircus | ENSCHIP00000007232 | 74.3243 | 62.1622 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 87.0748 | 76.8708 | panthera_leo | ENSPLOP00000011090 | 87.6712 | 77.3973 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 74.1497 | 61.9048 | bos_indicus_hybrid | ENSBIXP00005016093 | 73.6487 | 61.4865 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 67.3469 | 54.4218 | bos_indicus_hybrid | ENSBIXP00005015977 | 60 | 48.4848 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 84.3537 | 74.1497 | equus_caballus | ENSECAP00000073812 | 84.9315 | 74.6575 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 87.0748 | 77.551 | panthera_pardus | ENSPPRP00000028120 | 87.6712 | 78.0822 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 87.7551 | 78.9116 | marmota_marmota_marmota | ENSMMMP00000016938 | 88.3562 | 79.4521 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 76.1905 | 64.6258 | balaenoptera_musculus | ENSBMSP00010012151 | 63.2768 | 53.6723 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 85.034 | 75.5102 | ursus_americanus | ENSUAMP00000015106 | 85.6164 | 76.0274 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 78.2313 | 62.585 | monodelphis_domestica | ENSMODP00000008643 | 68.0473 | 54.4379 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 74.8299 | 63.9456 | bison_bison_bison | ENSBBBP00000016210 | 74.3243 | 63.5135 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 70.068 | 56.4626 | bison_bison_bison | ENSBBBP00000001778 | 70.068 | 56.4626 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 73.4694 | 60.5442 | bison_bison_bison | ENSBBBP00000018162 | 72.973 | 60.1351 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 74.8299 | 63.9456 | bison_bison_bison | ENSBBBP00000001776 | 75.8621 | 64.8276 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 77.551 | 64.6258 | monodon_monoceros | ENSMMNP00015013283 | 78.6207 | 65.5172 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 78.9116 | 68.7075 | sus_scrofa | ENSSSCP00000037223 | 78.9116 | 68.7075 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 74.1497 | 61.9048 | ovis_aries_rambouillet | ENSOARP00020017231 | 73.6487 | 61.4865 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 77.551 | 64.6258 | delphinapterus_leucas | ENSDLEP00000002301 | 50.6667 | 42.2222 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 72.1088 | 59.8639 | physeter_catodon | ENSPCTP00005003927 | 74.1259 | 61.5385 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 75.5102 | 63.2653 | mus_musculus | ENSMUSP00000159551 | 76.5517 | 64.1379 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 76.1905 | 64.6258 | mus_musculus | ENSMUSP00000154100 | 77.2414 | 65.5172 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 72.7891 | 62.585 | mus_musculus | ENSMUSP00000073525 | 74.3056 | 63.8889 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 84.3537 | 72.7891 | mus_musculus | ENSMUSP00000067434 | 85.5172 | 73.7931 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 68.0272 | 57.1429 | mus_musculus | ENSMUSP00000159552 | 68.0272 | 57.1429 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 76.1905 | 65.9864 | mesocricetus_auratus | ENSMAUP00000009246 | 77.2414 | 66.8966 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 73.4694 | 61.2245 | phocoena_sinus | ENSPSNP00000016888 | 74.4828 | 62.069 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 77.551 | 62.585 | camelus_dromedarius | ENSCDRP00005002541 | 81.4286 | 65.7143 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 87.0748 | 77.551 | panthera_tigris_altaica | ENSPTIP00000001602 | 87.6712 | 78.0822 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 82.9932 | 72.1088 | rattus_norvegicus | ENSRNOP00000034383 | 73.0539 | 63.4731 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 55.7823 | 49.6599 | prolemur_simus | ENSPSMP00000017459 | 83.6735 | 74.4898 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 81.6327 | 67.3469 | microtus_ochrogaster | ENSMOCP00000000790 | 82.1918 | 67.8082 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 76.8708 | 65.3061 | peromyscus_maniculatus_bairdii | ENSPEMP00000012976 | 67.6647 | 57.485 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 80.9524 | 70.068 | peromyscus_maniculatus_bairdii | ENSPEMP00000030293 | 81.5069 | 70.5479 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 78.2313 | 65.3061 | peromyscus_maniculatus_bairdii | ENSPEMP00000030309 | 77.1812 | 64.4295 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 75.5102 | 66.6667 | peromyscus_maniculatus_bairdii | ENSPEMP00000029513 | 76.0274 | 67.1233 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 78.9116 | 68.0272 | peromyscus_maniculatus_bairdii | ENSPEMP00000034475 | 79.4521 | 68.4931 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 74.8299 | 64.6258 | bos_mutus | ENSBMUP00000015916 | 74.3243 | 64.1892 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 70.068 | 55.102 | bos_mutus | ENSBMUP00000015925 | 70.068 | 55.102 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 73.4694 | 60.5442 | bos_mutus | ENSBMUP00000025800 | 73.9726 | 60.9589 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 73.4694 | 61.2245 | mus_spicilegus | ENSMSIP00000000265 | 73.9726 | 61.6438 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 84.3537 | 72.7891 | mus_spicilegus | ENSMSIP00000023087 | 85.5172 | 73.7931 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 74.8299 | 63.9456 | mus_spicilegus | ENSMSIP00000002601 | 76.3889 | 65.2778 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 73.4694 | 60.5442 | mus_spicilegus | ENSMSIP00000010568 | 56.8421 | 46.8421 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 75.5102 | 64.6258 | mus_spicilegus | ENSMSIP00000023154 | 76.5517 | 65.5172 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 76.8708 | 65.9864 | cricetulus_griseus_chok1gshd | ENSCGRP00001005193 | 53.0516 | 45.5399 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 78.9116 | 64.6258 | pteropus_vampyrus | ENSPVAP00000012796 | 79.4521 | 65.0685 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 78.2313 | 67.3469 | myotis_lucifugus | ENSMLUP00000019182 | 79.8611 | 68.75 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 83.6735 | 68.0272 | myotis_lucifugus | ENSMLUP00000016276 | 84.2466 | 68.4931 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 77.551 | 63.9456 | myotis_lucifugus | ENSMLUP00000017913 | 78.0822 | 64.3836 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 84.3537 | 67.3469 | myotis_lucifugus | ENSMLUP00000016273 | 84.9315 | 67.8082 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 81.6327 | 70.068 | myotis_lucifugus | ENSMLUP00000022677 | 81.6327 | 70.068 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 79.5918 | 67.3469 | myotis_lucifugus | ENSMLUP00000016811 | 81.25 | 68.75 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 82.3129 | 68.0272 | myotis_lucifugus | ENSMLUP00000021655 | 82.8767 | 68.4931 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 81.6327 | 69.3878 | rhinolophus_ferrumequinum | ENSRFEP00010025661 | 78.9474 | 67.1053 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 60.5442 | 51.7007 | neovison_vison | ENSNVIP00000017053 | 78.0702 | 66.6667 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 70.068 | 54.4218 | bos_taurus | ENSBTAP00000049727 | 70.068 | 54.4218 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 67.3469 | 54.4218 | bos_taurus | ENSBTAP00000070495 | 60 | 48.4848 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 72.7891 | 60.5442 | bos_taurus | ENSBTAP00000043945 | 63.6905 | 52.9762 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 84.3537 | 75.5102 | sciurus_vulgaris | ENSSVLP00005002095 | 85.5172 | 76.5517 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 65.3061 | 53.0612 | procavia_capensis | ENSPCAP00000005599 | 69.5652 | 56.5217 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 85.7143 | 76.8708 | nannospalax_galili | ENSNGAP00000018283 | 86.3014 | 77.3973 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 61.9048 | 53.0612 | cervus_hanglu_yarkandensis | ENSCHYP00000021581 | 50 | 42.8571 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 72.7891 | 59.8639 | cervus_hanglu_yarkandensis | ENSCHYP00000021249 | 64.8485 | 53.3333 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 78.2313 | 65.9864 | catagonus_wagneri | ENSCWAP00000008451 | 79.8611 | 67.3611 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 73.4694 | 61.9048 | otolemur_garnettii | ENSOGAP00000016686 | 73.9726 | 62.3288 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 88.4354 | 79.5918 | urocitellus_parryii | ENSUPAP00010002273 | 89.0411 | 80.137 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 70.7483 | 59.1837 | moschus_moschiferus | ENSMMSP00000011216 | 68.4211 | 57.2368 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 81.6327 | 69.3878 | mus_pahari | MGP_PahariEiJ_P0084619 | 82.7586 | 70.3448 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 81.6327 | 69.3878 | mus_pahari | MGP_PahariEiJ_P0084621 | 82.7586 | 70.3448 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 87.0748 | 77.551 | propithecus_coquereli | ENSPCOP00000004889 | 87.6712 | 78.0822 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 49.6599 | 38.7755 | chrysemys_picta_bellii | ENSCPBP00000012515 | 51.049 | 39.8601 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 57.8231 | 45.5782 | chrysemys_picta_bellii | ENSCPBP00000000807 | 53.7975 | 42.4051 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 57.1429 | 46.9388 | laticauda_laticaudata | ENSLLTP00000005473 | 50 | 41.0714 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 49.6599 | 38.7755 | pseudonaja_textilis | ENSPTXP00000024090 | 50.6944 | 39.5833 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 53.7415 | 40.8163 | podarcis_muralis | ENSPMRP00000034074 | 58.9552 | 44.7761 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 50.3401 | 37.415 | podarcis_muralis | ENSPMRP00000025114 | 59.2 | 44 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 49.6599 | 37.415 | podarcis_muralis | ENSPMRP00000034080 | 54.0741 | 40.7407 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 57.1429 | 42.8571 | podarcis_muralis | ENSPMRP00000034086 | 57.5342 | 43.1507 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 55.102 | 42.8571 | pelodiscus_sinensis | ENSPSIP00000000734 | 50.3106 | 39.1304 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 40.1361 | 32.6531 | gopherus_evgoodei | ENSGEVP00005021691 | 48.3607 | 39.3443 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 59.8639 | 48.2993 | gopherus_evgoodei | ENSGEVP00005021172 | 54.6584 | 44.0994 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 57.8231 | 46.9388 | chelonoidis_abingdonii | ENSCABP00000018574 | 54.8387 | 44.5161 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 52.381 | 38.7755 | terrapene_carolina_triunguis | ENSTMTP00000008306 | 53.8462 | 39.8601 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 57.1429 | 44.2177 | terrapene_carolina_triunguis | ENSTMTP00000008669 | 53.1646 | 41.1392 |
| ENSG00000214274 | homo_sapiens | ENSP00000381077 | 55.7823 | 42.1769 | paramormyrops_kingsleyae | ENSPKIP00000012614 | 36.7713 | 27.8027 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0001525 | involved_in angiogenesis | 4 | IBA,IDA,IMP,TAS | Homo_sapiens(9606) | Process |
| GO:0001541 | involved_in ovarian follicle development | 1 | NAS | Homo_sapiens(9606) | Process |
| GO:0001556 | involved_in oocyte maturation | 1 | NAS | Homo_sapiens(9606) | Process |
| GO:0001666 | involved_in response to hypoxia | 2 | IDA,NAS | Homo_sapiens(9606) | Process |
| GO:0001890 | involved_in placenta development | 1 | NAS | Homo_sapiens(9606) | Process |
| GO:0001938 | involved_in positive regulation of endothelial cell proliferation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0003677 | enables DNA binding | 1 | IC | Homo_sapiens(9606) | Function |
| GO:0003779 | enables actin binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0004519 | enables endonuclease activity | 1 | TAS | Homo_sapiens(9606) | Function |
| GO:0004521 | enables RNA endonuclease activity | 1 | TAS | Homo_sapiens(9606) | Function |
| GO:0004540 | enables RNA nuclease activity | 2 | IBA,IDA | Homo_sapiens(9606) | Function |
| GO:0005102 | enables signaling receptor binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0005507 | enables copper ion binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005576 | located_in extracellular region | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0005604 | located_in basement membrane | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005615 | is_active_in extracellular space | 2 | IBA,IDA | Homo_sapiens(9606) | Component |
| GO:0005634 | located_in nucleus | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005694 | located_in chromosome | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005730 | located_in nucleolus | 2 | IDA,ISS | Homo_sapiens(9606) | Component |
| GO:0005829 | located_in cytosol | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0006651 | involved_in diacylglycerol biosynthetic process | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0007154 | involved_in cell communication | 1 | NAS | Homo_sapiens(9606) | Process |
| GO:0007202 | involved_in activation of phospholipase C activity | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0008201 | enables heparin binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0009303 | involved_in rRNA transcription | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0009725 | involved_in response to hormone | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0015629 | located_in actin cytoskeleton | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0016078 | involved_in tRNA catabolic process | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0016477 | involved_in cell migration | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0017148 | involved_in negative regulation of translation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0019731 | involved_in antibacterial humoral response | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0019843 | enables rRNA binding | 1 | TAS | Homo_sapiens(9606) | Function |
| GO:0030041 | involved_in actin filament polymerization | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0030426 | located_in growth cone | 1 | ISS | Homo_sapiens(9606) | Component |
| GO:0031410 | located_in cytoplasmic vesicle | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0032148 | involved_in activation of protein kinase B activity | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0032311 | part_of angiogenin-PRI complex | 2 | IDA,IPI | Homo_sapiens(9606) | Component |
| GO:0032431 | involved_in activation of phospholipase A2 activity | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0042277 | enables peptide binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0042327 | involved_in positive regulation of phosphorylation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0042592 | involved_in homeostatic process | 1 | NAS | Homo_sapiens(9606) | Process |
| GO:0042803 | enables protein homodimerization activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0043025 | located_in neuronal cell body | 1 | ISS | Homo_sapiens(9606) | Component |
| GO:0045087 | involved_in innate immune response | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0048662 | involved_in negative regulation of smooth muscle cell proliferation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0050714 | involved_in positive regulation of protein secretion | 2 | IDA,ISS | Homo_sapiens(9606) | Process |
| GO:0050830 | involved_in defense response to Gram-positive bacterium | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0061844 | involved_in antimicrobial humoral immune response mediated by antimicrobial peptide | 1 | IBA | Homo_sapiens(9606) | Process |