Welcome to
RBP World!
Description
| Ensembl ID | ENSG00000196664 | Gene ID | 51284 | Accession | 15631 |
| Symbol | TLR7 | Alias | IMD74;SLEB17;TLR7-like | Full Name | toll like receptor 7 |
| Status | Confidence | Length | 23290 bases | Strand | Plus strand |
| Position | X : 12867072 - 12890361 | RNA binding domain | N.A. | ||
| Summary | The protein encoded by this gene is a member of the Toll-like receptor (TLR) family which plays a fundamental role in pathogen recognition and activation of innate immunity. TLRs are highly conserved from Drosophila to humans and share structural and functional similarities. The human TLR family comprises 11 members. They recognize pathogen-associated molecular patterns (PAMPs) that are expressed on infectious agents, and mediate the production of cytokines necessary for the development of effective immunity. For the recognition of structural components in foreign microorganisms, the various TLRs exhibit different patterns of expression as well; in this way for example, TLR-3, -7, and -8 are essential in the recognition of single-stranded RNA viruses. TLR7 senses single-stranded RNA oligonucleotides containing guanosine- and uridine-rich sequences from RNA viruses, a recognition occuring in the endosomes of plasmacytoid dendritic cells and B cells. This gene is predominantly expressed in lung, placenta, and spleen, and is phylogenetically related and lies in close proximity to another family member, TLR8, on chromosome X. [provided by RefSeq, Aug 2020] | ||||
RNA binding domains (RBDs)
| Protein | Domain | Pfam ID | E-value | Domain number |
|---|
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|---|---|---|---|
| 34868046 | Anti-TLR7 Antibody Protects Against Lupus Nephritis in NZBWF1 Mice by Targeting B Cells and Patrolling Monocytes. | Yusuke Murakami | 2021-01-01 | Frontiers in immunology |
| 26683146 | Endosomal Toll-like receptors in clinically overt and silent autoimmunity. | M Robert Clancy | 2016-01-01 | Immunological reviews |
| 27329272 | CD4 T cell epitope specificity determines follicular versus non-follicular helper differentiation in the polyclonal response to influenza infection or vaccination. | G A Zackery Knowlden | 2016-06-22 | Scientific reports |
| 29500721 | Innate Immune Signaling and Alcohol Use Disorders. | G Leon Coleman | 2018-01-01 | Handbook of experimental pharmacology |
| 29439290 | Review: Cell Death, Nucleic Acids, and Immunity: Inflammation Beyond the Grave. | B Keith Elkon | 2018-06-01 | Arthritis & rheumatology (Hoboken, N.J.) |
| 37513775 | The Potential of Probiotics as Ingestible Adjuvants and Immune Modulators for Antiviral Immunity and Management of SARS-CoV-2 Infection and COVID-19. | Sophie Tomkinson | 2023-07-11 | Pathogens (Basel, Switzerland) |
| 21389263 | Neutrophils activate plasmacytoid dendritic cells by releasing self-DNA-peptide complexes in systemic lupus erythematosus. | Roberto Lande | 2011-03-09 | Science translational medicine |
| 19633559 | Innate immunity and inflammation in systemic sclerosis. | Robert Lafyatis | 2009-11-01 | Current opinion in rheumatology |
| 19592080 | Toll-like receptor 7 and 9 defects in common variable immunodeficiency. | E Joyce Yu | 2009-08-01 | The Journal of allergy and clinical immunology |
| 34393859 | SIGIRR and TNFAIP3 Are Differentially Expressed in Both PBMC and TNF-α Secreting Cells of Patients With Major Depressive Disorder. | Chin-Chuen Lin | 2021-01-01 | Frontiers in psychiatry |
| 23151919 | Therapeutic applications of nucleic acids and their analogues in Toll-like receptor signaling. | Vijayakumar Gosu | 2012-11-14 | Molecules (Basel, Switzerland) |
| 26302811 | Innate and Adaptive Immune Responses in Chronic HCV Infection. | B Lynn Dustin | 2017-01-01 | Current drug targets |
| 28471483 | Bone Marrow-Derived Mesenchymal Stem Cells From Patients With Systemic Lupus Erythematosus Have a Senescence-Associated Secretory Phenotype Mediated by a Mitochondrial Antiviral Signaling Protein-Interferon-β Feedback Loop. | Lin Gao | 2017-08-01 | Arthritis & rheumatology (Hoboken, N.J.) |
| 24740301 | Interleukin-7 and Toll-like receptor 7 induce synergistic B cell and T cell activation. | Angela Bikker | 2014-01-01 | PloS one |
| 21389264 | Netting neutrophils are major inducers of type I IFN production in pediatric systemic lupus erythematosus. | S Gina Garcia-Romo | 2011-03-09 | Science translational medicine |
| 25010440 | Advances in understanding the role of type I interferons in systemic lupus erythematosus. | K Mary Crow | 2014-09-01 | Current opinion in rheumatology |
| 26442097 | Polymorphisms in Toll-like receptor genes are associated with vitiligo. | Tanel Traks | 2015-01-01 | Frontiers in genetics |
| 35456913 | STAT1 and Its Crucial Role in the Control of Viral Infections. | Manlio Tolomeo | 2022-04-07 | International journal of molecular sciences |
| 26545385 | Human TLR8 senses UR/URR motifs in bacterial and mitochondrial RNA. | Anne Krüger | 2015-12-01 | EMBO reports |
| 28604502 | Association of HIV-1 Gag-Specific IgG Antibodies With Natural Control of HIV-1 Infection in Individuals Not Carrying HLA-B*57: 01 Is Only Observed in Viremic Controllers. | Christian M Tjiam | 2017-11-01 | Journal of acquired immune deficiency syndromes (1999) |
| 27412723 | An efficient method for gene silencing in human primary plasmacytoid dendritic cells: silencing of the TLR7/IRF-7 pathway as a proof of concept. | Nikaïa Smith | 2016-07-14 | Scientific reports |
| 29181003 | The Human-Associated Archaeon Methanosphaera stadtmanae Is Recognized through Its RNA and Induces TLR8-Dependent NLRP3 Inflammasome Activation. | Tim Vierbuchen | 2017-01-01 | Frontiers in immunology |
| 19006693 | IRAK-4- and MyD88-dependent pathways are essential for the removal of developing autoreactive B cells in humans. | Isabelle Isnardi | 2008-11-14 | Immunity |
| 16330816 | Immune stimulation mediated by autoantigen binding sites within small nuclear RNAs involves Toll-like receptors 7 and 8. | Jörg Vollmer | 2005-12-05 | The Journal of experimental medicine |
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| TLR7-201 | ENST00000380659 | 5003 | 1049aa | ENSP00000370034 |
| TLR7-202 | ENST00000484204 | 299 | No protein | - |
Phenotypes
| ensgID | Trait | pValue | Pubmed ID |
|---|
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
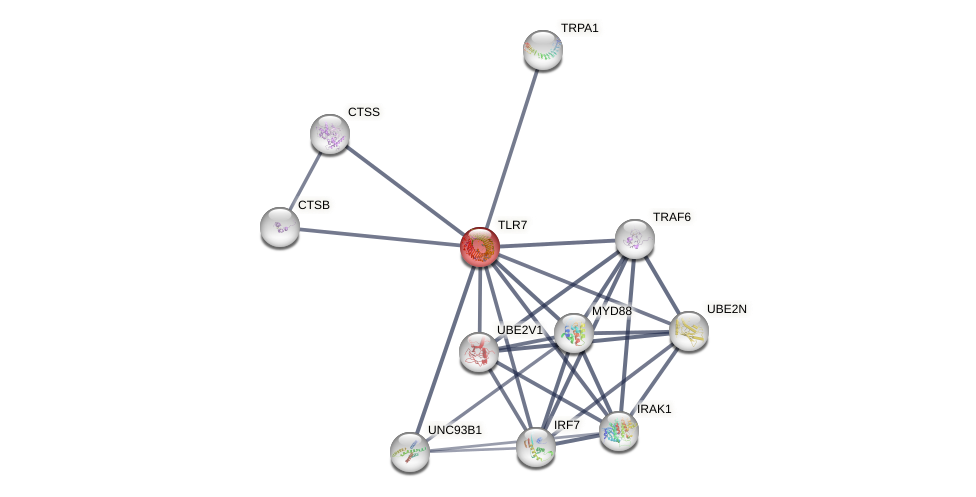
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000196664 | homo_sapiens | ENSP00000306864 | 28.9747 | 17.682 | homo_sapiens | ENSP00000370034 | 18.5891 | 11.3441 |
| ENSG00000196664 | homo_sapiens | ENSP00000215539 | 39.5041 | 25.6198 | homo_sapiens | ENSP00000370034 | 22.7836 | 14.776 |
| ENSG00000196664 | homo_sapiens | ENSP00000363089 | 39.2133 | 23.0036 | homo_sapiens | ENSP00000370034 | 31.3632 | 18.3985 |
| ENSG00000196664 | homo_sapiens | ENSP00000344242 | 38.5488 | 21.542 | homo_sapiens | ENSP00000370034 | 16.2059 | 9.05624 |
| ENSG00000196664 | homo_sapiens | ENSP00000218032 | 59.9424 | 42.3631 | homo_sapiens | ENSP00000370034 | 59.4852 | 42.04 |
| ENSG00000196664 | homo_sapiens | ENSP00000291592 | 38.1323 | 23.3463 | homo_sapiens | ENSP00000370034 | 9.34223 | 5.71973 |
| ENSG00000196664 | homo_sapiens | ENSP00000367157 | 38.1818 | 21.8182 | homo_sapiens | ENSP00000370034 | 10.0095 | 5.71973 |
| ENSG00000196664 | homo_sapiens | ENSP00000379880 | 38.2239 | 21.6216 | homo_sapiens | ENSP00000370034 | 9.43756 | 5.33842 |
| ENSG00000196664 | homo_sapiens | ENSP00000328625 | 34.9711 | 18.9306 | homo_sapiens | ENSP00000370034 | 23.0696 | 12.4881 |
| ENSG00000196664 | homo_sapiens | ENSP00000308925 | 36.7448 | 19.3588 | homo_sapiens | ENSP00000370034 | 28.408 | 14.9666 |
| ENSG00000196664 | homo_sapiens | ENSP00000296795 | 41.3717 | 24.8894 | homo_sapiens | ENSP00000370034 | 35.653 | 21.449 |
| ENSG00000196664 | homo_sapiens | ENSP00000496355 | 39.7436 | 22.6107 | homo_sapiens | ENSP00000370034 | 32.5071 | 18.4938 |
| ENSG00000196664 | homo_sapiens | ENSP00000332668 | 35.1275 | 22.6629 | homo_sapiens | ENSP00000370034 | 11.8208 | 7.62631 |
| ENSG00000196664 | homo_sapiens | ENSP00000298386 | 32.7586 | 18.435 | homo_sapiens | ENSP00000370034 | 23.5462 | 13.2507 |
| ENSG00000196664 | homo_sapiens | ENSP00000494425 | 38.1378 | 22.3214 | homo_sapiens | ENSP00000370034 | 28.5033 | 16.6826 |
| ENSG00000196664 | homo_sapiens | ENSP00000260061 | 37.7644 | 23.565 | homo_sapiens | ENSP00000370034 | 23.8322 | 14.8713 |
| ENSG00000196664 | homo_sapiens | ENSP00000354932 | 38.0407 | 20.6107 | homo_sapiens | ENSP00000370034 | 28.5033 | 15.4433 |
| ENSG00000196664 | homo_sapiens | ENSP00000361185 | 31.0909 | 15.8182 | homo_sapiens | ENSP00000370034 | 16.3012 | 8.29361 |
| ENSG00000196664 | homo_sapiens | ENSP00000303248 | 32.6288 | 17.8336 | homo_sapiens | ENSP00000370034 | 23.5462 | 12.8694 |
| ENSG00000196664 | homo_sapiens | ENSP00000294635 | 11.7081 | 6.49559 | homo_sapiens | ENSP00000370034 | 13.918 | 7.72164 |
| ENSG00000196664 | homo_sapiens | ENSP00000424718 | 35.9296 | 19.3467 | homo_sapiens | ENSP00000370034 | 27.2641 | 14.6806 |
| ENSG00000196664 | homo_sapiens | ENSP00000256447 | 39.1831 | 22.3903 | homo_sapiens | ENSP00000370034 | 24.6902 | 14.1087 |
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 99.428 | 98.5701 | nomascus_leucogenys | ENSNLEP00000011104 | 99.428 | 98.5701 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 99.714 | 99.6187 | pan_paniscus | ENSPPAP00000009572 | 99.714 | 99.6187 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 99.428 | 98.9514 | pongo_abelii | ENSPPYP00000038399 | 99.428 | 98.9514 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 99.5234 | 99.428 | pan_troglodytes | ENSPTRP00000047494 | 99.5234 | 99.428 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 99.714 | 99.428 | gorilla_gorilla | ENSGGOP00000020624 | 99.714 | 99.428 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 84.2707 | 70.0667 | ornithorhynchus_anatinus | ENSOANP00000004155 | 78.6477 | 65.3915 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 83.5081 | 72.6406 | sarcophilus_harrisii | ENSSHAP00000038315 | 83.4286 | 72.5714 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 84.1754 | 73.0219 | notamacropus_eugenii | ENSMEUP00000013829 | 83.9354 | 72.8137 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 77.693 | 71.4967 | dasypus_novemcinctus | ENSDNOP00000019813 | 89.9558 | 82.7815 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 83.2221 | 75.8818 | erinaceus_europaeus | ENSEEUP00000002924 | 83.4608 | 76.0994 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 86.654 | 78.837 | echinops_telfairi | ENSETEP00000004208 | 86.654 | 78.837 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 96.6635 | 93.9943 | callithrix_jacchus | ENSCJAP00000007128 | 96.6635 | 93.9943 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 98.8561 | 97.8074 | cercocebus_atys | ENSCATP00000042815 | 98.8561 | 97.8074 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 98.9514 | 97.9028 | macaca_fascicularis | ENSMFAP00000056275 | 98.9514 | 97.9028 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 98.9514 | 97.9028 | macaca_mulatta | ENSMMUP00000018719 | 98.9514 | 97.9028 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 98.9514 | 97.9028 | macaca_nemestrina | ENSMNEP00000027405 | 98.9514 | 97.9028 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 98.9514 | 97.9028 | papio_anubis | ENSPANP00000008761 | 98.9514 | 97.9028 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 98.7607 | 97.7121 | mandrillus_leucophaeus | ENSMLEP00000033999 | 98.7607 | 97.7121 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 92.755 | 86.1773 | tursiops_truncatus | ENSTTRP00000003025 | 92.6667 | 86.0952 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 92.469 | 85.4147 | vulpes_vulpes | ENSVVUP00000026246 | 86.5299 | 79.9286 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 89.3232 | 80.8389 | mus_spretus | MGP_SPRETEiJ_P0097162 | 89.2381 | 80.7619 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 92.3737 | 86.2726 | equus_asinus | ENSEASP00005060369 | 91.9355 | 85.8634 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 92.0877 | 86.368 | capra_hircus | ENSCHIP00000023443 | 91.6509 | 85.9583 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 90.6578 | 84.1754 | loxodonta_africana | ENSLAFP00000028694 | 91.4423 | 84.9038 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 92.469 | 86.1773 | panthera_leo | ENSPLOP00000029273 | 92.381 | 86.0952 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 92.8503 | 85.796 | ailuropoda_melanoleuca | ENSAMEP00000020832 | 92.7619 | 85.7143 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 91.7064 | 86.368 | bos_indicus_hybrid | ENSBIXP00005029635 | 91.2713 | 85.9583 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 92.3737 | 86.2726 | equus_caballus | ENSECAP00000064301 | 92.3737 | 86.2726 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 92.469 | 85.51 | canis_lupus_familiaris | ENSCAFP00845039166 | 86.5299 | 80.0178 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 92.469 | 86.082 | panthera_pardus | ENSPPRP00000021798 | 92.381 | 86 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 91.8017 | 84.8427 | marmota_marmota_marmota | ENSMMMP00000004774 | 91.9771 | 85.0048 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 92.6597 | 86.368 | balaenoptera_musculus | ENSBMSP00010026127 | 92.5714 | 86.2857 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 90.4671 | 83.3174 | cavia_porcellus | ENSCPOP00000028818 | 90.4671 | 83.3174 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 92.3737 | 86.2726 | felis_catus | ENSFCAP00000039674 | 90.4762 | 84.5005 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 92.5643 | 85.51 | ursus_americanus | ENSUAMP00000035160 | 91.6903 | 84.7026 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 57.8646 | 54.9094 | bison_bison_bison | ENSBBBP00000002603 | 57.8646 | 54.9094 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 92.5643 | 85.9867 | monodon_monoceros | ENSMMNP00015021444 | 92.4762 | 85.9048 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 92.6597 | 85.0334 | sus_scrofa | ENSSSCP00000012898 | 92.2201 | 84.63 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 93.041 | 87.7026 | microcebus_murinus | ENSMICP00000048998 | 92.9524 | 87.619 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 90.3718 | 82.8408 | octodon_degus | ENSODEP00000004822 | 90.3718 | 82.8408 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 91.5157 | 83.0315 | jaculus_jaculus | ENSJJAP00000020387 | 91.4286 | 82.9524 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 92.6597 | 85.51 | ursus_maritimus | ENSUMAP00000006805 | 92.5714 | 85.4286 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 67.9695 | 61.1058 | ochotona_princeps | ENSOPRP00000015519 | 68.2297 | 61.3397 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 92.3737 | 85.796 | delphinapterus_leucas | ENSDLEP00000027613 | 91.9355 | 85.389 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 92.9457 | 86.1773 | physeter_catodon | ENSPCTP00005021070 | 91.3777 | 84.7235 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 89.4185 | 81.7922 | mus_caroli | MGP_CAROLIEiJ_P0093392 | 89.3333 | 81.7143 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 89.2278 | 80.7436 | mus_musculus | ENSMUSP00000107787 | 89.1429 | 80.6667 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 89.6092 | 80.8389 | dipodomys_ordii | ENSDORP00000026642 | 89.5238 | 80.7619 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 91.897 | 83.9847 | mustela_putorius_furo | ENSMPUP00000002534 | 89.012 | 81.3481 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 96.6635 | 94.4709 | aotus_nancymaae | ENSANAP00000026575 | 96.6635 | 94.4709 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 96.3775 | 94.2803 | saimiri_boliviensis_boliviensis | ENSSBOP00000003105 | 96.3775 | 94.2803 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 65.7769 | 61.1058 | vicugna_pacos | ENSVPAP00000000301 | 65.9026 | 61.2225 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 98.7607 | 97.6168 | chlorocebus_sabaeus | ENSCSAP00000019028 | 98.7607 | 97.6168 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 88.7512 | 79.7903 | mesocricetus_auratus | ENSMAUP00000019401 | 88.6667 | 79.7143 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 92.469 | 86.082 | phocoena_sinus | ENSPSNP00000021726 | 92.381 | 86 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 91.8017 | 85.0334 | camelus_dromedarius | ENSCDRP00005014589 | 85.0707 | 78.7986 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 93.8036 | 88.4652 | carlito_syrichta | ENSTSYP00000019414 | 93.7143 | 88.381 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 92.3737 | 85.8913 | panthera_tigris_altaica | ENSPTIP00000019983 | 92.2857 | 85.8095 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 88.7512 | 80.5529 | rattus_norvegicus | ENSRNOP00000094649 | 87.1723 | 79.1199 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 93.7083 | 87.8932 | prolemur_simus | ENSPSMP00000037418 | 93.619 | 87.8095 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 88.1792 | 79.3136 | microtus_ochrogaster | ENSMOCP00000011105 | 88.0952 | 79.2381 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 89.5138 | 80.1716 | peromyscus_maniculatus_bairdii | ENSPEMP00000000923 | 89.4286 | 80.0952 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 91.7064 | 86.368 | bos_mutus | ENSBMUP00000019534 | 91.2713 | 85.9583 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 88.9418 | 79.7903 | cricetulus_griseus_chok1gshd | ENSCGRP00001015821 | 88.8571 | 79.7143 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 91.6111 | 86.082 | pteropus_vampyrus | ENSPVAP00000008906 | 91.6111 | 86.082 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 91.5157 | 84.1754 | myotis_lucifugus | ENSMLUP00000019342 | 91.6031 | 84.2557 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 84.5567 | 73.3079 | vombatus_ursinus | ENSVURP00010029268 | 82.9747 | 71.9364 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 92.8503 | 87.2259 | rhinolophus_ferrumequinum | ENSRFEP00010025176 | 92.8503 | 87.2259 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 91.5157 | 83.6034 | neovison_vison | ENSNVIP00000025533 | 91.7782 | 83.8432 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 91.7064 | 86.368 | bos_taurus | ENSBTAP00000071792 | 91.2713 | 85.9583 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 92.0877 | 85.0334 | heterocephalus_glaber_female | ENSHGLP00000000964 | 92.0877 | 85.0334 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 99.2374 | 98.0934 | rhinopithecus_bieti | ENSRBIP00000004854 | 99.2374 | 98.0934 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 92.469 | 85.51 | canis_lupus_dingo | ENSCAFP00020003258 | 91.5094 | 84.6226 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 92.183 | 86.654 | sciurus_vulgaris | ENSSVLP00005027150 | 92.3591 | 86.8195 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 83.6988 | 73.0219 | phascolarctos_cinereus | ENSPCIP00000040748 | 82.1328 | 71.6558 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 39.2755 | 35.653 | procavia_capensis | ENSPCAP00000009024 | 71.6522 | 65.0435 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 89.3232 | 80.9342 | nannospalax_galili | ENSNGAP00000023836 | 89.2381 | 80.8571 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 91.6111 | 86.2726 | bos_grunniens | ENSBGRP00000030589 | 91.1765 | 85.8634 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 91.3251 | 84.0801 | chinchilla_lanigera | ENSCLAP00000000459 | 91.3251 | 84.0801 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 92.0877 | 86.368 | cervus_hanglu_yarkandensis | ENSCHYP00000035954 | 92 | 86.2857 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 92.3737 | 85.4147 | catagonus_wagneri | ENSCWAP00000023793 | 92.2857 | 85.3333 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 91.7064 | 84.8427 | ictidomys_tridecemlineatus | ENSSTOP00000020078 | 91.1848 | 84.3602 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 91.9924 | 85.51 | otolemur_garnettii | ENSOGAP00000014402 | 92.0802 | 85.5916 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 96.5682 | 94.1849 | cebus_imitator | ENSCCAP00000011708 | 96.5682 | 94.1849 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 91.8017 | 84.8427 | urocitellus_parryii | ENSUPAP00010002829 | 91.9771 | 85.0048 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 91.9924 | 86.1773 | moschus_moschiferus | ENSMMSP00000015767 | 91.9048 | 86.0952 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 99.3327 | 98.1888 | rhinopithecus_roxellana | ENSRROP00000004175 | 99.3327 | 98.1888 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 89.2278 | 81.3155 | mus_pahari | MGP_PahariEiJ_P0090993 | 89.1429 | 81.2381 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 51.2869 | 33.1745 | petromyzon_marinus | ENSPMAP00000006887 | 56.871 | 36.7865 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 56.0534 | 39.2755 | petromyzon_marinus | ENSPMAP00000002547 | 55.2113 | 38.6854 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 45.6625 | 31.9352 | eptatretus_burgeri | ENSEBUP00000003579 | 57.8502 | 40.4589 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 73.4032 | 57.0067 | callorhinchus_milii | ENSCMIP00000021859 | 73.9673 | 57.4448 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 76.3584 | 60.3432 | latimeria_chalumnae | ENSLACP00000001158 | 75.566 | 59.717 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 75.4052 | 60.3432 | latimeria_chalumnae | ENSLACP00000004758 | 76.5731 | 61.2778 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 73.4032 | 57.8646 | lepisosteus_oculatus | ENSLOCP00000012197 | 72.9167 | 57.4811 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 67.3022 | 52.0496 | clupea_harengus | ENSCHAP00000061091 | 69.5566 | 53.7931 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 72.2593 | 55.3861 | danio_rerio | ENSDARP00000105671 | 72.5359 | 55.5981 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 72.164 | 56.7207 | carassius_auratus | ENSCARP00000005322 | 72.164 | 56.7207 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 72.8313 | 57.102 | carassius_auratus | ENSCARP00000059802 | 72.4171 | 56.7773 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 73.2126 | 56.4347 | astyanax_mexicanus | ENSAMXP00000026765 | 72.5898 | 55.9546 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 71.4967 | 55.1001 | esox_lucius | ENSELUP00000067316 | 71.09 | 54.7867 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 70.5434 | 55.1954 | electrophorus_electricus | ENSEEEP00000026825 | 71.775 | 56.1591 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 73.4032 | 56.6254 | oncorhynchus_kisutch | ENSOKIP00005017591 | 72.7101 | 56.0907 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 73.2126 | 56.1487 | oncorhynchus_mykiss | ENSOMYP00000060949 | 73.2126 | 56.1487 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 73.4986 | 56.53 | salmo_salar | ENSSSAP00000035772 | 72.5306 | 55.7855 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 73.4986 | 56.53 | salmo_trutta | ENSSTUP00000031989 | 72.5306 | 55.7855 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 72.6406 | 55.5767 | gadus_morhua | ENSGMOP00000032968 | 71.8868 | 55 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 72.2593 | 55.4814 | gadus_morhua | ENSGMOP00000024016 | 71.442 | 54.8539 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 71.592 | 55.3861 | fundulus_heteroclitus | ENSFHEP00000033866 | 71.1848 | 55.0711 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 72.164 | 55.1954 | poecilia_reticulata | ENSPREP00000002624 | 71.9582 | 55.038 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 71.878 | 55.2908 | xiphophorus_maculatus | ENSXMAP00000004436 | 71.7412 | 55.1855 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 72.2593 | 56.6254 | oryzias_latipes | ENSORLP00000031133 | 71.9848 | 56.4103 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 73.7846 | 56.816 | cyclopterus_lumpus | ENSCLMP00005032126 | 73.4345 | 56.5465 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 73.7846 | 56.816 | oreochromis_niloticus | ENSONIP00000025981 | 73.5043 | 56.6002 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 73.6892 | 56.7207 | haplochromis_burtoni | ENSHBUP00000034814 | 72.3783 | 55.7116 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 73.5939 | 56.7207 | astatotilapia_calliptera | ENSACLP00000009792 | 72.2846 | 55.7116 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 71.9733 | 54.5281 | sparus_aurata | ENSSAUP00010036842 | 69.9722 | 53.012 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 73.4986 | 57.102 | lates_calcarifer | ENSLCAP00010013353 | 73.639 | 57.2111 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 44.4233 | 35.081 | lates_calcarifer | ENSLCAP00010049506 | 72.6989 | 57.4103 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 76.3584 | 60.7245 | xenopus_tropicalis | ENSXETP00000011725 | 75.9962 | 60.4364 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 82.1735 | 68.5415 | chrysemys_picta_bellii | ENSCPBP00000017267 | 81.3975 | 67.8942 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 81.1249 | 66.5396 | crocodylus_porosus | ENSCPRP00005008862 | 79.1628 | 64.9302 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 70.0667 | 55.1954 | laticauda_laticaudata | ENSLLTP00000003642 | 72.1295 | 56.8204 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 69.3994 | 54.4328 | notechis_scutatus | ENSNSUP00000010828 | 71.7241 | 56.2562 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 69.7807 | 54.7188 | pseudonaja_textilis | ENSPTXP00000007869 | 70.793 | 55.5126 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 79.123 | 65.3956 | anas_platyrhynchos_platyrhynchos | ENSAPLP00000003630 | 76.2167 | 62.9936 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 78.3603 | 63.775 | gallus_gallus | ENSGALP00010002615 | 77.6204 | 63.1728 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 78.3603 | 63.9657 | meleagris_gallopavo | ENSMGAP00000015580 | 78.51 | 64.0879 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 78.9323 | 65.205 | serinus_canaria | ENSSCAP00000018507 | 79.0831 | 65.3295 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 76.8351 | 63.2984 | serinus_canaria | ENSSCAP00000018512 | 80.6 | 66.4 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 79.5043 | 65.0143 | parus_major | ENSPMJP00000015192 | 76.5138 | 62.5688 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 15.1573 | 12.2021 | parus_major | ENSPMJP00000015191 | 79.8995 | 64.3216 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 74.7378 | 57.2927 | anolis_carolinensis | ENSACAP00000001544 | 74.2424 | 56.9129 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 73.8799 | 57.102 | neolamprologus_brichardi | ENSNBRP00000011317 | 73.5992 | 56.8851 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 66.1582 | 52.0496 | naja_naja | ENSNNAP00000011515 | 70.7441 | 55.6575 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 73.6892 | 57.2927 | podarcis_muralis | ENSPMRP00000011122 | 74.686 | 58.0676 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 78.1697 | 64.4423 | geospiza_fortis | ENSGFOP00000005543 | 79.3037 | 65.3772 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 78.6463 | 64.8236 | geospiza_fortis | ENSGFOP00000020219 | 79.2507 | 65.3218 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 22.5929 | 18.5891 | geospiza_fortis | ENSGFOP00000020222 | 80.339 | 66.1017 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 78.7417 | 64.8236 | geospiza_fortis | ENSGFOP00000020216 | 78.8921 | 64.9475 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 78.9323 | 64.7283 | taeniopygia_guttata | ENSTGUP00000032154 | 79.0831 | 64.852 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 73.0219 | 55.1001 | erpetoichthys_calabaricus | ENSECRP00000017310 | 72.3324 | 54.5798 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 72.2593 | 56.6254 | kryptolebias_marmoratus | ENSKMAP00000023686 | 71.1737 | 55.7746 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 70.9247 | 55.4814 | salvator_merianae | ENSSMRP00000029353 | 71.1962 | 55.6938 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 72.8313 | 56.244 | oryzias_javanicus | ENSOJAP00000026724 | 72.5546 | 56.0304 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 73.3079 | 56.0534 | amphilophus_citrinellus | ENSACIP00000010847 | 72.891 | 55.7346 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 73.8799 | 57.8646 | dicentrarchus_labrax | ENSDLAP00005060498 | 73.5992 | 57.6448 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 73.2126 | 55.8627 | amphiprion_percula | ENSAPEP00000033915 | 72.9345 | 55.6505 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 72.6406 | 56.7207 | oryzias_sinensis | ENSOSIP00000025936 | 72.3647 | 56.5052 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 81.1249 | 67.8742 | pelodiscus_sinensis | ENSPSIP00000000410 | 81.0476 | 67.8095 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 82.0782 | 68.3508 | gopherus_evgoodei | ENSGEVP00005008827 | 82 | 68.2857 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 76.7398 | 64.633 | chelonoidis_abingdonii | ENSCABP00000015485 | 81.8922 | 68.9725 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 73.0219 | 56.244 | seriola_dumerili | ENSSDUP00000004349 | 72.8137 | 56.0836 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 73.6892 | 56.3394 | cottoperca_gobio | ENSCGOP00000050215 | 73.2701 | 56.019 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 79.7903 | 65.3956 | struthio_camelus_australis | ENSSCUP00000022268 | 75.4734 | 61.8575 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 79.2183 | 65.1096 | anser_brachyrhynchus | ENSABRP00000011904 | 78.175 | 64.2521 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 72.9266 | 55.7674 | gasterosteus_aculeatus | ENSGACP00000005259 | 73.4165 | 56.142 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 72.8313 | 54.9094 | cynoglossus_semilaevis | ENSCSEP00000015651 | 72.1435 | 54.3909 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 72.3546 | 55.3861 | poecilia_formosa | ENSPFOP00000025729 | 71.6038 | 54.8113 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 73.5939 | 57.1973 | sander_lucioperca | ENSSLUP00000000609 | 73.1754 | 56.872 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 79.7903 | 65.1096 | strigops_habroptila | ENSSHBP00005022836 | 79.9427 | 65.234 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 73.4986 | 56.1487 | amphiprion_ocellaris | ENSAOCP00000012889 | 73.2194 | 55.9354 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 77.3117 | 65.205 | terrapene_carolina_triunguis | ENSTMTP00000019046 | 82.1682 | 69.3009 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 77.4071 | 65.3003 | terrapene_carolina_triunguis | ENSTMTP00000003329 | 82.2695 | 69.4022 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 73.2126 | 56.244 | hucho_hucho | ENSHHUP00000041035 | 72.2484 | 55.5033 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 72.7359 | 56.7207 | betta_splendens | ENSBSLP00000040980 | 72.5975 | 56.6128 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 73.2126 | 56.816 | pygocentrus_nattereri | ENSPNAP00000025476 | 71.7757 | 55.7009 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 74.1659 | 56.9113 | anabas_testudineus | ENSATEP00000019327 | 73.8841 | 56.6952 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 73.4032 | 56.4347 | seriola_lalandi_dorsalis | ENSSLDP00000001130 | 73.1939 | 56.2738 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 73.4986 | 55.5767 | labrus_bergylta | ENSLBEP00000026932 | 73.2194 | 55.3656 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 73.6892 | 56.9113 | pundamilia_nyererei | ENSPNYP00000015309 | 73.4093 | 56.6952 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 73.4986 | 56.0534 | mastacembelus_armatus | ENSMAMP00000019868 | 73.2194 | 55.8405 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 72.6406 | 55.5767 | stegastes_partitus | ENSSPAP00000010498 | 72.3647 | 55.3656 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 73.4032 | 56.6254 | oncorhynchus_tshawytscha | ENSOTSP00005060935 | 72.7101 | 56.0907 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 73.4032 | 56.6254 | oncorhynchus_tshawytscha | ENSOTSP00005068428 | 72.7101 | 56.0907 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 73.1173 | 55.7674 | acanthochromis_polyacanthus | ENSAPOP00000033418 | 72.8395 | 55.5556 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 76.7398 | 62.8217 | aquila_chrysaetos_chrysaetos | ENSACCP00020013640 | 80.9045 | 66.2312 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 72.6406 | 57.102 | nothobranchius_furzeri | ENSNFUP00015019223 | 71.9547 | 56.5628 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 71.7827 | 55.1954 | cyprinodon_variegatus | ENSCVAP00000027218 | 72.3343 | 55.6196 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 71.9733 | 54.8141 | denticeps_clupeoides | ENSDCDP00000034413 | 71.6319 | 54.5541 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 73.4032 | 54.8141 | larimichthys_crocea | ENSLCRP00005025934 | 73.1244 | 54.6059 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 72.7359 | 55.3861 | takifugu_rubripes | ENSTRUP00000020082 | 72.8749 | 55.4919 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 14.204 | 11.9161 | ficedula_albicollis | ENSFALP00000020132 | 65.9292 | 55.3097 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 14.2993 | 11.5348 | ficedula_albicollis | ENSFALP00000032773 | 67.5676 | 54.5045 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 78.6463 | 64.061 | ficedula_albicollis | ENSFALP00000004661 | 78.7966 | 64.1834 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 71.6873 | 53.8608 | hippocampus_comes | ENSHCOP00000002794 | 70.9434 | 53.3019 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 72.6406 | 57.2927 | cyprinus_carpio_carpio | ENSCCRP00000005032 | 72.6406 | 57.2927 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 78.4557 | 65.0143 | coturnix_japonica | ENSCJPP00005022916 | 78.6055 | 65.1385 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 72.2593 | 55.3861 | poecilia_latipinna | ENSPLAP00000018153 | 72.0532 | 55.2281 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 72.45 | 57.1973 | sinocyclocheilus_grahami | ENSSGRP00000102147 | 72.45 | 57.1973 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 72.9266 | 57.4833 | sinocyclocheilus_grahami | ENSSGRP00000071860 | 72.7878 | 57.3739 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 72.8313 | 55.6721 | tetraodon_nigroviridis | ENSTNIP00000016967 | 72.9008 | 55.7252 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 73.4986 | 56.6254 | paramormyrops_kingsleyae | ENSPKIP00000031114 | 72.9423 | 56.1968 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 72.45 | 56.6254 | scleropages_formosus | ENSSFOP00015004856 | 71.7658 | 56.0907 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 74.0705 | 58.3413 | leptobrachium_leishanense | ENSLLEP00000005761 | 73.9296 | 58.2303 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 66.8255 | 48.7131 | scophthalmus_maximus | ENSSMAP00000068793 | 45.5787 | 33.225 |
| ENSG00000196664 | homo_sapiens | ENSP00000370034 | 72.6406 | 56.6254 | oryzias_melastigma | ENSOMEP00000005763 | 72.3647 | 56.4103 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0000139 | located_in Golgi membrane | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0001932 | involved_in regulation of protein phosphorylation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0002224 | involved_in toll-like receptor signaling pathway | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0003725 | enables double-stranded RNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0003727 | enables single-stranded RNA binding | 2 | IMP,ISS | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005737 | located_in cytoplasm | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005764 | located_in lysosome | 1 | ISS | Homo_sapiens(9606) | Component |
| GO:0005768 | located_in endosome | 1 | ISS | Homo_sapiens(9606) | Component |
| GO:0005783 | located_in endoplasmic reticulum | 1 | ISS | Homo_sapiens(9606) | Component |
| GO:0005789 | located_in endoplasmic reticulum membrane | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0005886 | is_active_in plasma membrane | 2 | IBA,IDA | Homo_sapiens(9606) | Component |
| GO:0006954 | involved_in inflammatory response | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0007249 | involved_in I-kappaB kinase/NF-kappaB signaling | 2 | IBA,IMP | Homo_sapiens(9606) | Process |
| GO:0007252 | involved_in I-kappaB phosphorylation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0007254 | involved_in JNK cascade | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0010008 | located_in endosome membrane | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0032009 | located_in early phagosome | 1 | ISS | Homo_sapiens(9606) | Component |
| GO:0032722 | involved_in positive regulation of chemokine production | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0032727 | involved_in positive regulation of interferon-alpha production | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0032728 | involved_in positive regulation of interferon-beta production | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0032729 | involved_in positive regulation of type II interferon production | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0032755 | involved_in positive regulation of interleukin-6 production | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0032757 | involved_in positive regulation of interleukin-8 production | 2 | IDA,IMP | Homo_sapiens(9606) | Process |
| GO:0034154 | involved_in toll-like receptor 7 signaling pathway | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0035197 | enables siRNA binding | 1 | ISS | Homo_sapiens(9606) | Function |
| GO:0036020 | located_in endolysosome membrane | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0038187 | enables pattern recognition receptor activity | 1 | IBA | Homo_sapiens(9606) | Function |
| GO:0043235 | part_of receptor complex | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0045087 | involved_in innate immune response | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0045944 | involved_in positive regulation of transcription by RNA polymerase II | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0050729 | involved_in positive regulation of inflammatory response | 1 | IC | Homo_sapiens(9606) | Process |
| GO:0051607 | involved_in defense response to virus | 3 | IBA,IEP,IMP | Homo_sapiens(9606) | Process |
| GO:0060907 | involved_in positive regulation of macrophage cytokine production | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0070305 | involved_in response to cGMP | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0071260 | involved_in cellular response to mechanical stimulus | 1 | IEP | Homo_sapiens(9606) | Process |
| GO:0098586 | involved_in cellular response to virus | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:1901224 | involved_in positive regulation of NIK/NF-kappaB signaling | 1 | IDA | Homo_sapiens(9606) | Process |