RBP Type
Canonical_RBPs
Diseases
Drug
N.A.
Main interacting RNAs
ncRNA
Moonlighting functions
N.A.
Localizations
N.A.
BulkPerturb-seq
N.A.
Ensembl ID ENSG00000181381 Gene ID
91351 Accession
26429
Symbol
DDX60L
Alias
NA
Full Name
DExD/H-box 60 like
Status
Confidence
Length
123780 bases
Strand
Minus strand
Position
4 : 168356735 - 168480514
RNA binding domain
ResIII , DEAD
Summary
This gene encodes a member of the DExD/H-box helicase family of proteins, a subset of the super family 2 helicases. Members of the DExD/H-box helicase family share a conserved functional core comprised of two RecA-like globular domains. These domains contain conserved motifs that mediate ATP binding, ATP hydrolysis, nucleic acid binding, and RNA unwinding. In addition to functions in RNA metabolism, members of this family are involved in anti-viral immunity and act as cytosolic sensors of viral nucleic acids. The protein encoded by this gene has been shown to inhibit hepatitis C virus replication in response to interferon stimulation in cell culture. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Sep 2016]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
ENSP00000422202
DEAD
PF00270 1.4e-14
1
ENSP00000422202
ResIII
PF04851 3.1e-10
2
ENSP00000422423
DEAD
PF00270 2e-14
1
ENSP00000422423
ResIII
PF04851 6.8e-10
2
ENSP00000507872
DEAD
PF00270 2e-14
1
ENSP00000507872
ResIII
PF04851 6.8e-10
2
ENSP00000421026
DEAD
PF00270 3.5e-06
1
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
Name
Transcript ID
bp
Protein
Translation ID
DDX60L-207
ENST00000510590
3509
No protein
-
DDX60L-216
ENST00000682922
6759
1706aa
ENSP00000507872
DDX60L-208
ENST00000511577
6781
1706aa
ENSP00000422423
DDX60L-211
ENST00000513103
507
No protein
-
DDX60L-206
ENST00000505890
4568
1308aa
ENSP00000422202
DDX60L-213
ENST00000514580
813
195aa
ENSP00000422920
DDX60L-201
ENST00000503190
562
No protein
-
DDX60L-205
ENST00000505863
2605
868aa
ENSP00000421026
DDX60L-202
ENST00000504793
549
No protein
-
DDX60L-204
ENST00000505696
588
124aa
ENSP00000424310
DDX60L-214
ENST00000514748
632
98aa
ENSP00000424396
DDX60L-215
ENST00000515088
2652
No protein
-
DDX60L-212
ENST00000513901
623
No protein
-
DDX60L-209
ENST00000512371
428
77aa
ENSP00000427386
DDX60L-203
ENST00000505150
655
No protein
-
DDX60L-210
ENST00000512958
769
No protein
-
Pathway ID
Pathway Name
Source
ensgID
Trait
pValue
Pubmed ID
26755 Blood Pressure 8.5170000E-005 - 59967 Hypertension 2.8164309E-005 - 64277 Liver Cirrhosis 6E-7 20708005 64278 Non-alcoholic Fatty Liver Disease 6E-7 20708005 84032 Energy Intake 8E-6 23251661
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
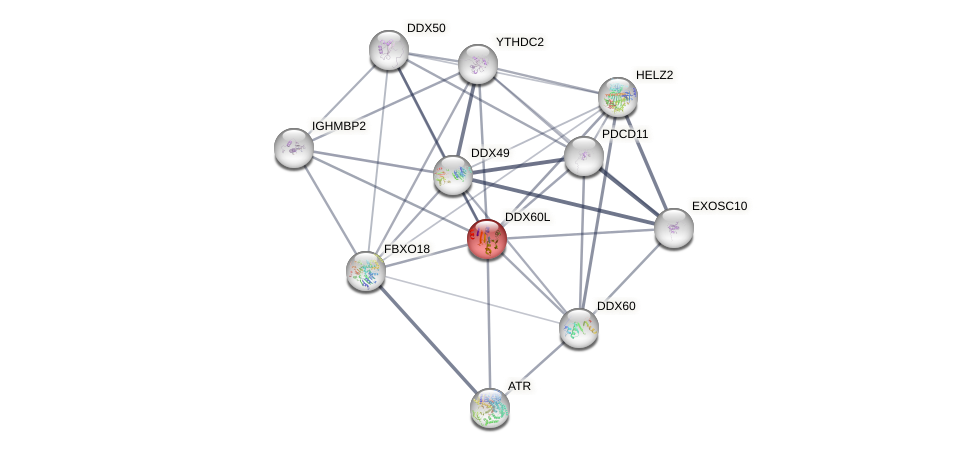
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
ENSG00000181381 homo_sapiens ENSP00000264233 12.8571 6.67954 homo_sapiens ENSP00000507872 19.5193 10.1407 ENSG00000181381 homo_sapiens ENSP00000364543 26.7255 15.0883 homo_sapiens ENSP00000507872 19.5193 11.0199 ENSG00000181381 homo_sapiens ENSP00000317123 17.5094 8.2397 homo_sapiens ENSP00000507872 21.9226 10.3165 ENSG00000181381 homo_sapiens ENSP00000359454 22.0209 10.1742 homo_sapiens ENSP00000507872 18.5229 8.55803 ENSG00000181381 homo_sapiens ENSP00000295488 30.881 15.8038 homo_sapiens ENSP00000507872 19.9297 10.1993 ENSG00000181381 homo_sapiens ENSP00000377344 78.8551 65.479 homo_sapiens ENSP00000507872 79.1325 65.7093 ENSG00000181381 homo_sapiens ENSP00000358159 17.9837 8.03815 homo_sapiens ENSP00000507872 23.2122 10.3751 ENSG00000181381 homo_sapiens ENSP00000230640 26.6795 14.2994 homo_sapiens ENSP00000507872 16.2954 8.73388
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
ENSG00000181381 homo_sapiens ENSP00000507872 97.5381 95.7796 nomascus_leucogenys ENSNLEP00000007985 97.481 95.7235 ENSG00000181381 homo_sapiens ENSP00000507872 99.0035 98.2415 pan_paniscus ENSPPAP00000003493 98.484 97.7259 ENSG00000181381 homo_sapiens ENSP00000507872 97.6553 96.0727 pongo_abelii ENSPPYP00000016967 97.6553 96.0727 ENSG00000181381 homo_sapiens ENSP00000507872 99.0035 98.3001 pan_troglodytes ENSPTRP00000075463 98.484 97.7843 ENSG00000181381 homo_sapiens ENSP00000507872 98.476 97.5967 gorilla_gorilla ENSGGOP00000014436 97.9592 97.0845 ENSG00000181381 homo_sapiens ENSP00000507872 89.3904 83.177 callithrix_jacchus ENSCJAP00000008692 91.4817 85.123 ENSG00000181381 homo_sapiens ENSP00000507872 95.3107 91.7937 cercocebus_atys ENSCATP00000043419 94.9766 91.472 ENSG00000181381 homo_sapiens ENSP00000507872 95.1934 91.6178 macaca_fascicularis ENSMFAP00000028009 95.1934 91.6178 ENSG00000181381 homo_sapiens ENSP00000507872 95.3693 91.6764 macaca_mulatta ENSMMUP00000046407 94.7583 91.0891 ENSG00000181381 homo_sapiens ENSP00000507872 95.4865 91.9695 macaca_nemestrina ENSMNEP00000010322 95.0408 91.5403 ENSG00000181381 homo_sapiens ENSP00000507872 94.7831 91.442 papio_anubis ENSPANP00000012037 95.0059 91.6569 ENSG00000181381 homo_sapiens ENSP00000507872 95.0176 91.5006 mandrillus_leucophaeus ENSMLEP00000006982 94.7399 91.2332 ENSG00000181381 homo_sapiens ENSP00000507872 84.4666 75.1465 equus_asinus ENSEASP00005031883 83.8278 74.5782 ENSG00000181381 homo_sapiens ENSP00000507872 26.8464 22.9191 loxodonta_africana ENSLAFP00000024539 78.4247 66.9521 ENSG00000181381 homo_sapiens ENSP00000507872 84.5252 75.4396 equus_caballus ENSECAP00000042990 83.9348 74.9127 ENSG00000181381 homo_sapiens ENSP00000507872 15.9437 7.32708 marmota_marmota_marmota ENSMMMP00000008711 26.9041 12.364 ENSG00000181381 homo_sapiens ENSP00000507872 88.5698 83.0012 aotus_nancymaae ENSANAP00000029321 92.078 86.2888 ENSG00000181381 homo_sapiens ENSP00000507872 90.973 85.4631 saimiri_boliviensis_boliviensis ENSSBOP00000036488 90.7602 85.2632 ENSG00000181381 homo_sapiens ENSP00000507872 95.0176 91.2075 chlorocebus_sabaeus ENSCSAP00000014893 94.6846 90.8878 ENSG00000181381 homo_sapiens ENSP00000507872 84.7597 74.7948 carlito_syrichta ENSTSYP00000012080 84.6109 74.6635 ENSG00000181381 homo_sapiens ENSP00000507872 93.3177 89.5076 rhinopithecus_bieti ENSRBIP00000006022 94.3128 90.4621 ENSG00000181381 homo_sapiens ENSP00000507872 90.2696 84.5252 cebus_imitator ENSCCAP00000024775 91.0165 85.2246 ENSG00000181381 homo_sapiens ENSP00000507872 95.5451 92.0281 rhinopithecus_roxellana ENSRROP00000031998 95.1547 91.6521 ENSG00000181381 homo_sapiens ENSP00000507872 20.9261 9.61313 caenorhabditis_elegans Y46G5A.4.1 16.6434 7.64569 ENSG00000181381 homo_sapiens ENSP00000507872 1.0551 0.410317 oncorhynchus_mykiss ENSOMYP00000000359 17.3077 6.73077 ENSG00000181381 homo_sapiens ENSP00000507872 1.4068 0.410317 pygocentrus_nattereri ENSPNAP00000029897 21.6216 6.30631 ENSG00000181381 homo_sapiens ENSP00000507872 15.2989 6.6823 saccharomyces_cerevisiae YGL251C 21.9882 9.60404 ENSG00000181381 homo_sapiens ENSP00000507872 19.9883 8.85111 saccharomyces_cerevisiae YER172C 15.7651 6.98104 ENSG00000181381 homo_sapiens ENSP00000507872 5.39273 2.8136 poecilia_latipinna ENSPLAP00000000676 28.5714 14.9068 ENSG00000181381 homo_sapiens ENSP00000507872 4.33763 2.34467 sinocyclocheilus_grahami ENSSGRP00000075535 22.7692 12.3077
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0003724 enables RNA helicase activity 1 IEA Homo_sapiens(9606) Function GO:0003725 enables double-stranded RNA binding 1 IBA Homo_sapiens(9606) Function GO:0003727 enables single-stranded RNA binding 1 IBA Homo_sapiens(9606) Function GO:0005524 enables ATP binding 1 IEA Homo_sapiens(9606) Function GO:0005737 is_active_in cytoplasm 1 IBA Homo_sapiens(9606) Component GO:0016887 enables ATP hydrolysis activity 1 IEA Homo_sapiens(9606) Function GO:0051607 involved_in defense response to virus 1 IBA Homo_sapiens(9606) Process
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.