Welcome to
RBP World!
Functions and Diseases
| RBP Type | Canonical_RBPs |
| Diseases | CancerCHIKVSARS-COV-OC43HIVIAVRVSARS-COV-2SINVDengueZika |
| Drug | N.A. |
| Main interacting RNAs | mRNA |
| Moonlighting functions | N.A. |
| Localizations | Stress granucleP-body |
| BulkPerturb-seq | DataSet_01_121 DataSet_01_288 |
Description
| Ensembl ID | ENSG00000169564 | Gene ID | 5093 | Accession | 8647 |
| Symbol | PCBP1 | Alias | HNRPX;HNRPE1;hnRNP-X;HEL-S-85;hnRNP-E1 | Full Name | poly(rC) binding protein 1 |
| Status | Confidence | Length | 1727 bases | Strand | Plus strand |
| Position | 2 : 70087477 - 70089203 | RNA binding domain | KH_1 | ||
| Summary | This intronless gene is thought to have been generated by retrotransposition of a fully processed PCBP-2 mRNA. This gene and PCBP-2 have paralogues (PCBP3 and PCBP4) which are thought to have arisen as a result of duplication events of entire genes. The protein encoded by this gene appears to be multifunctional. It along with PCBP-2 and hnRNPK corresponds to the major cellular poly(rC)-binding protein. It contains three K-homologous (KH) domains which may be involved in RNA binding. This encoded protein together with PCBP-2 also functions as translational coactivators of poliovirus RNA via a sequence-specific interaction with stem-loop IV of the IRES and promote poliovirus RNA replication by binding to its 5'-terminal cloverleaf structure. It has also been implicated in translational control of the 15-lipoxygenase mRNA, human Papillomavirus type 16 L2 mRNA, and hepatitis A virus RNA. The encoded protein is also suggested to play a part in formation of a sequence-specific alpha-globin mRNP complex which is associated with alpha-globin mRNA stability. [provided by RefSeq, Jul 2008] | ||||
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|---|---|---|---|
| 28076324 | Poly(C)-binding protein 1 mediates drug resistance in colorectal cancer. | Jiani Guo | 2017-02-21 | Oncotarget |
| 36902478 | The Functions of TRIM56 in Antiviral Innate Immunity and Tumorigenesis. | Lin Fu | 2023-03-06 | International journal of molecular sciences |
| 29138420 | Expression of both poly r(C) binding protein 1 (PCBP1) and miRNA-3978 is suppressed in peritoneal gastric cancer metastasis. | Fu-Jian Ji | 2017-11-14 | Scientific reports |
| 15246275 | A posttranscriptional regulator of Kaposi's sarcoma-associated herpesvirus interacts with RNA-binding protein PCBP1 and controls gene expression through the IRES. | Ken Nishimura | 2004-08-01 | Virology |
| 27911100 | Mitochondria and Iron: current questions. | T Bibbin Paul | 2017-01-01 | Expert review of hematology |
| 35907982 | The RNA-binding protein PCBP1 represses lung adenocarcinoma progression by stabilizing DKK1 mRNA and subsequently downregulating β-catenin. | Yujia Zheng | 2022-07-30 | Journal of translational medicine |
| 19843848 | Nuclear receptor coregulators in cancer biology. | W Bert O'Malley | 2009-11-01 | Cancer research |
| 25559010 | Role of heme in cardiovascular physiology and disease. | Teodor Konrad Sawicki | 2015-01-05 | Journal of the American Heart Association |
| 32922440 | Poly C Binding Protein 1 Regulates p62/SQSTM1 mRNA Stability and Autophagic Degradation to Repress Tumor Progression. | Wenliang Zhang | 2020-01-01 | Frontiers in genetics |
| 19754138 | Iron uptake and transport in plants: the good, the bad, and the ionome. | Joe Morrissey | 2009-10-01 | Chemical reviews |
| 32835748 | Management versus miscues in the cytosolic labile iron pool: The varied functions of iron chaperones. | C Caroline Philpott | 2020-11-01 | Biochimica et biophysica acta. Molecular cell research |
| 35290903 | Poly(rC)-binding protein 1 represses ferritinophagy-mediated ferroptosis in head and neck cancer. | Jaewang Lee | 2022-05-01 | Redox biology |
| 30247716 | Enterovirus 71 antagonizes the inhibition of the host intrinsic antiviral factor A3G. | Zhaolong Li | 2018-11-30 | Nucleic acids research |
| 31326364 | Two ways of escaping from oxidative RNA damage: Selective degradation and cell death. | Takashi Ishii | 2019-09-01 | DNA repair |
| 33416185 | miRNA‑490‑3p promotes the metastatic progression of invasive ductal carcinoma. | Ning Lu | 2021-02-01 | Oncology reports |
| 37426488 | Functional Role of RBP in Osteosarcoma: Regulatory Mechanism and Clinical Therapy. | Ziyuan Que | 2023-01-01 | Analytical cellular pathology (Amsterdam) |
| 32647012 | PCBP1 and PCBP2 both bind heavily oxidized RNA but cause opposing outcomes, suppressing or increasing apoptosis under oxidative conditions. | Takashi Ishii | 2020-08-21 | The Journal of biological chemistry |
| 35795237 | PCBP-1 Regulates the Transcription and Alternative Splicing of Inflammation and Ubiquitination-Related Genes in PC12 Cell. | Aishanjiang Yusufujiang | 2022-01-01 | Frontiers in aging neuroscience |
| 33579665 | Biomarkers of nucleic acid oxidation - A summary state-of-the-art. | Mu-Rong Chao | 2021-06-01 | Redox biology |
| 23823326 | Single-stranded DNA-binding proteins: multiple domains for multiple functions. | H Thayne Dickey | 2013-07-02 | Structure (London, England : 1993) |
| 27836661 | Phosphorylation of poly(rC) binding protein 1 (PCBP1) contributes to stabilization of mu opioid receptor (MOR) mRNA via interaction with AU-rich element RNA-binding protein 1 (AUF1) and poly A binding protein (PABP). | Kyu Cheol Hwang | 2017-01-20 | Gene |
| 36406269 | Dynamics and energetics of PCBP1 binding to severely oxidized RNA. | Natacha Gillet | 2022-01-01 | Frontiers in molecular biosciences |
| 18656221 | The linker domain of poly(rC) binding protein 2 is a major determinant in poliovirus cap-independent translation. | Polen Sean | 2008-09-01 | Virology |
| 31406370 | A PCBP1-BolA2 chaperone complex delivers iron for cytosolic [2Fe-2S] cluster assembly. | J Sarju Patel | 2019-09-01 | Nature chemical biology |
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| PCBP1-201 | ENST00000303577 | 1727 | 356aa | ENSP00000305556 |
Phenotypes
| ensgID | Trait | pValue | Pubmed ID |
|---|
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
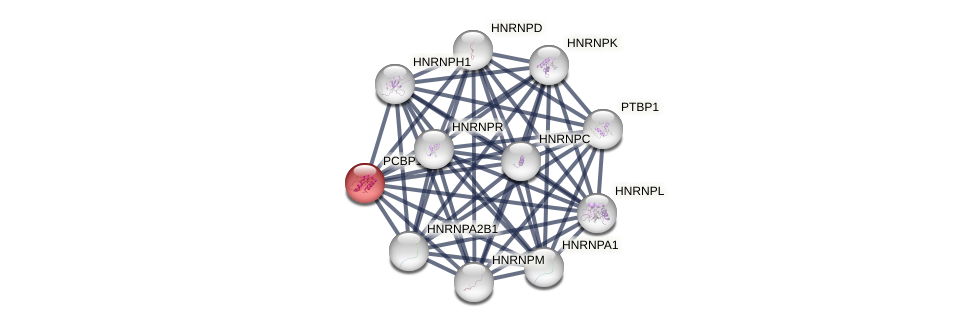
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000169564 | homo_sapiens | ENSP00000290341 | 26.1698 | 15.078 | homo_sapiens | ENSP00000305556 | 42.4157 | 24.4382 |
| ENSG00000169564 | homo_sapiens | ENSP00000417196 | 58.0645 | 45.4094 | homo_sapiens | ENSP00000305556 | 65.7303 | 51.4045 |
| ENSG00000169564 | homo_sapiens | ENSP00000263257 | 28.0488 | 16.2602 | homo_sapiens | ENSP00000305556 | 38.764 | 22.4719 |
| ENSG00000169564 | homo_sapiens | ENSP00000505796 | 73.0458 | 64.1509 | homo_sapiens | ENSP00000305556 | 76.1236 | 66.8539 |
| ENSG00000169564 | homo_sapiens | ENSP00000365439 | 30.819 | 20.0431 | homo_sapiens | ENSP00000305556 | 40.1685 | 26.1236 |
| ENSG00000169564 | homo_sapiens | ENSP00000448762 | 88.9503 | 82.8729 | homo_sapiens | ENSP00000305556 | 90.4494 | 84.2697 |
| ENSG00000169564 | homo_sapiens | ENSP00000371634 | 25.8765 | 15.192 | homo_sapiens | ENSP00000305556 | 43.5393 | 25.5618 |
| ENSG00000169564 | homo_sapiens | ENSP00000359804 | 22.205 | 13.9752 | homo_sapiens | ENSP00000305556 | 40.1685 | 25.2809 |
| ENSG00000169564 | homo_sapiens | ENSP00000258729 | 25.734 | 14.8532 | homo_sapiens | ENSP00000305556 | 41.8539 | 24.1573 |
| ENSG00000169564 | homo_sapiens | ENSP00000318177 | 26.049 | 16.0839 | homo_sapiens | ENSP00000305556 | 41.8539 | 25.8427 |
| ENSG00000169564 | homo_sapiens | ENSP00000438875 | 26.43 | 14.9901 | homo_sapiens | ENSP00000305556 | 37.6404 | 21.3483 |
| ENSG00000169564 | homo_sapiens | ENSP00000471146 | 20.3481 | 12.8514 | homo_sapiens | ENSP00000305556 | 42.6966 | 26.9663 |
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | nomascus_leucogenys | ENSNLEP00000022598 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 99.1573 | 98.8764 | pan_paniscus | ENSPPAP00000025989 | 98.0556 | 97.7778 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | pongo_abelii | ENSPPYP00000013749 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | pan_troglodytes | ENSPTRP00000020647 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | gorilla_gorilla | ENSGGOP00000023163 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 80.0562 | 79.4944 | dasypus_novemcinctus | ENSDNOP00000034664 | 99.6504 | 98.951 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 96.6292 | 94.6629 | callithrix_jacchus | ENSCJAP00000071168 | 98.5673 | 96.5616 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | callithrix_jacchus | ENSCJAP00000072017 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | cercocebus_atys | ENSCATP00000004155 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | macaca_fascicularis | ENSMFAP00000063749 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | macaca_mulatta | ENSMMUP00000046093 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | macaca_nemestrina | ENSMNEP00000000621 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 95.2247 | 95.2247 | papio_anubis | ENSPANP00000029646 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | mandrillus_leucophaeus | ENSMLEP00000006936 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 90.4494 | 90.4494 | vulpes_vulpes | ENSVVUP00000039348 | 98.773 | 98.773 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | mus_spretus | MGP_SPRETEiJ_P0077741 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | equus_asinus | ENSEASP00005056688 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | capra_hircus | ENSCHIP00000003735 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | panthera_leo | ENSPLOP00000016842 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | ailuropoda_melanoleuca | ENSAMEP00000016998 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 91.2921 | 91.2921 | bos_indicus_hybrid | ENSBIXP00005028269 | 99.0854 | 99.0854 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | oryctolagus_cuniculus | ENSOCUP00000020092 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | equus_caballus | ENSECAP00000074799 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 99.7191 | 99.7191 | canis_lupus_familiaris | ENSCAFP00845037555 | 99.7191 | 99.7191 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 91.0112 | 90.1685 | canis_lupus_familiaris | ENSCAFP00845032185 | 97.5904 | 96.6867 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | panthera_pardus | ENSPPRP00000028437 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | marmota_marmota_marmota | ENSMMMP00000005448 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | balaenoptera_musculus | ENSBMSP00010022255 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | cavia_porcellus | ENSCPOP00000012166 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | felis_catus | ENSFCAP00000017172 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | bison_bison_bison | ENSBBBP00000008895 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | monodon_monoceros | ENSMMNP00015020023 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 90.4494 | 90.4494 | sus_scrofa | ENSSSCP00000076382 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | microcebus_murinus | ENSMICP00000022546 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 85.9551 | 82.8652 | octodon_degus | ENSODEP00000000295 | 88.9535 | 85.7558 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | jaculus_jaculus | ENSJJAP00000000871 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | ovis_aries_rambouillet | ENSOARP00020007574 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | delphinapterus_leucas | ENSDLEP00000007261 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | physeter_catodon | ENSPCTP00005014669 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | mus_caroli | MGP_CAROLIEiJ_P0074795 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | mus_musculus | ENSMUSP00000054863 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | dipodomys_ordii | ENSDORP00000025060 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 97.191 | 97.191 | mustela_putorius_furo | ENSMPUP00000007117 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 76.9663 | 75.5618 | aotus_nancymaae | ENSANAP00000002964 | 96.8198 | 95.053 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 87.3596 | 85.6742 | aotus_nancymaae | ENSANAP00000031286 | 95.6923 | 93.8462 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 95.2247 | 94.382 | aotus_nancymaae | ENSANAP00000018293 | 96.5812 | 95.7265 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 76.6854 | 75.5618 | aotus_nancymaae | ENSANAP00000027753 | 96.4664 | 95.053 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 73.8764 | 73.0337 | aotus_nancymaae | ENSANAP00000008935 | 96.6912 | 95.5882 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 79.7753 | 77.809 | aotus_nancymaae | ENSANAP00000018546 | 95.6229 | 93.266 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 88.2022 | 87.9213 | aotus_nancymaae | ENSANAP00000016412 | 94.8641 | 94.5619 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 89.6067 | 88.4831 | aotus_nancymaae | ENSANAP00000011997 | 94.6588 | 93.4718 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 85.6742 | 83.427 | saimiri_boliviensis_boliviensis | ENSSBOP00000038488 | 89.4428 | 87.0968 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | chlorocebus_sabaeus | ENSCSAP00000018949 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 57.5843 | 56.7416 | tupaia_belangeri | ENSTBEP00000013625 | 57.7465 | 56.9014 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 99.7191 | 99.7191 | mesocricetus_auratus | ENSMAUP00000008419 | 99.7191 | 99.7191 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | phocoena_sinus | ENSPSNP00000026858 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 26.6854 | 25.2809 | camelus_dromedarius | ENSCDRP00005013539 | 87.156 | 82.5688 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 99.7191 | 99.4382 | carlito_syrichta | ENSTSYP00000031826 | 99.7191 | 99.4382 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | panthera_tigris_altaica | ENSPTIP00000004929 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | rattus_norvegicus | ENSRNOP00000092121 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 83.427 | 83.1461 | prolemur_simus | ENSPSMP00000025457 | 94.2857 | 93.9683 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | microtus_ochrogaster | ENSMOCP00000008163 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | peromyscus_maniculatus_bairdii | ENSPEMP00000029614 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 99.7191 | bos_mutus | ENSBMUP00000027719 | 100 | 99.7191 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 96.9101 | 96.9101 | mus_spicilegus | ENSMSIP00000017583 | 96.9101 | 96.9101 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | cricetulus_griseus_chok1gshd | ENSCGRP00001014090 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | myotis_lucifugus | ENSMLUP00000012890 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | rhinolophus_ferrumequinum | ENSRFEP00010022321 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 99.7191 | 99.7191 | neovison_vison | ENSNVIP00000004431 | 99.7191 | 99.7191 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 91.2921 | 91.2921 | bos_taurus | ENSBTAP00000049383 | 99.0854 | 99.0854 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 94.1011 | 91.2921 | heterocephalus_glaber_female | ENSHGLP00000028163 | 72.5108 | 70.3463 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | sciurus_vulgaris | ENSSVLP00005020164 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | nannospalax_galili | ENSNGAP00000013270 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | nannospalax_galili | ENSNGAP00000009210 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 96.6292 | 95.7865 | bos_grunniens | ENSBGRP00000008135 | 100 | 99.1279 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 99.4382 | 99.4382 | chinchilla_lanigera | ENSCLAP00000025783 | 99.4382 | 99.4382 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | cervus_hanglu_yarkandensis | ENSCHYP00000018032 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 99.7191 | 99.7191 | catagonus_wagneri | ENSCWAP00000004817 | 99.7191 | 99.7191 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | ictidomys_tridecemlineatus | ENSSTOP00000026830 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 97.191 | 97.191 | otolemur_garnettii | ENSOGAP00000018442 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 94.1011 | 91.0112 | cebus_imitator | ENSCCAP00000022814 | 92.5414 | 89.5028 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | cebus_imitator | ENSCCAP00000004600 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | urocitellus_parryii | ENSUPAP00010006802 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | moschus_moschiferus | ENSMMSP00000013586 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 99.1573 | 99.1573 | rhinopithecus_roxellana | ENSRROP00000002392 | 93.883 | 93.883 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 92.1348 | 92.1348 | rhinopithecus_roxellana | ENSRROP00000003137 | 98.7952 | 98.7952 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | mus_pahari | MGP_PahariEiJ_P0050906 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 100 | 100 | propithecus_coquereli | ENSPCOP00000028562 | 100 | 100 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 54.4944 | 37.3596 | caenorhabditis_elegans | Y119D3B.17.2 | 44.9074 | 30.787 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 26.9663 | 16.2921 | caenorhabditis_elegans | Y59A8B.10d.1 | 23.0216 | 13.9089 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 35.9551 | 19.382 | caenorhabditis_elegans | C36E6.1b.1 | 19.9688 | 10.7644 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 34.8315 | 18.2584 | caenorhabditis_elegans | M01A10.1.1 | 21.1604 | 11.0921 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 29.7753 | 15.1685 | caenorhabditis_elegans | M88.5c.1 | 12.4122 | 6.32318 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 35.1124 | 17.9775 | caenorhabditis_elegans | C12D8.1b.1 | 20.4583 | 10.4746 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 64.6067 | 48.5955 | drosophila_melanogaster | FBpp0293521 | 40.1396 | 30.192 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 35.1124 | 21.3483 | drosophila_melanogaster | FBpp0306419 | 16.0256 | 9.74359 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 34.8315 | 20.2247 | drosophila_melanogaster | FBpp0293680 | 19.4357 | 11.2853 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 36.236 | 19.6629 | drosophila_melanogaster | FBpp0428307 | 16.1857 | 8.78294 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 52.809 | 37.6404 | ciona_intestinalis | ENSCINP00000001984 | 52.6611 | 37.535 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 22.4719 | 11.7978 | ciona_intestinalis | ENSCINP00000025950 | 28.169 | 14.7887 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 38.2022 | 20.2247 | ciona_intestinalis | ENSCINP00000008726 | 23.0118 | 12.1827 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 25.2809 | 17.1348 | ciona_intestinalis | ENSCINP00000019569 | 28.3019 | 19.1824 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 25.8427 | 15.7303 | ciona_intestinalis | ENSCINP00000030435 | 34.981 | 21.2928 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 36.5169 | 20.5056 | astyanax_mexicanus | ENSAMXP00000028943 | 41.4013 | 23.2484 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 25.5618 | 17.6966 | ciona_savignyi | ENSCSAVP00000004755 | 43.3333 | 30 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 23.8764 | 18.2584 | ciona_savignyi | ENSCSAVP00000007214 | 69.6721 | 53.2787 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 38.2022 | 20.7865 | ciona_savignyi | ENSCSAVP00000018813 | 23.4483 | 12.7586 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 28.9326 | 18.8202 | ciona_savignyi | ENSCSAVP00000007375 | 33.3333 | 21.6828 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 31.1798 | 19.382 | ciona_savignyi | ENSCSAVP00000005505 | 20.5176 | 12.7542 |
| ENSG00000169564 | homo_sapiens | ENSP00000305556 | 16.0112 | 8.98876 | ciona_savignyi | ENSCSAVP00000005450 | 16.8639 | 9.46746 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0000981 | enables DNA-binding transcription factor activity, RNA polymerase II-specific | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0003697 | enables single-stranded DNA binding | 2 | IBA,IDA | Homo_sapiens(9606) | Function |
| GO:0003723 | enables RNA binding | 2 | HDA,IDA | Homo_sapiens(9606) | Function |
| GO:0003729 | enables mRNA binding | 2 | IBA,IDA | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005634 | is_active_in nucleus | 1 | IBA | Homo_sapiens(9606) | Component |
| GO:0005654 | is_active_in nucleoplasm | 3 | IBA,IDA,TAS | Homo_sapiens(9606) | Component |
| GO:0005737 | is_active_in cytoplasm | 2 | IBA,IDA | Homo_sapiens(9606) | Component |
| GO:0005829 | located_in cytosol | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0014069 | located_in postsynaptic density | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0016020 | located_in membrane | 1 | HDA | Homo_sapiens(9606) | Component |
| GO:0016607 | located_in nuclear speck | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0036464 | located_in cytoplasmic ribonucleoprotein granule | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0039694 | involved_in viral RNA genome replication | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0045296 | enables cadherin binding | 1 | HDA | Homo_sapiens(9606) | Function |
| GO:0045944 | involved_in positive regulation of transcription by RNA polymerase II | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0070062 | located_in extracellular exosome | 1 | HDA | Homo_sapiens(9606) | Component |
| GO:0098847 | enables sequence-specific single stranded DNA binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:1990904 | part_of ribonucleoprotein complex | 1 | IEA | Homo_sapiens(9606) | Component |