Welcome to
RBP World!
Description
| Ensembl ID | ENSG00000167986 | Gene ID | 1642 | Accession | 2717 |
| Symbol | DDB1 | Alias | XPE;DDBA;XAP1;XPCE;XPE-BF;UV-DDB1;WHIKERS | Full Name | damage specific DNA binding protein 1 |
| Status | Confidence | Length | 43146 bases | Strand | Minus strand |
| Position | 11 : 61299451 - 61342596 | RNA binding domain | CPSF_A | ||
| Summary | The protein encoded by this gene is the large subunit (p127) of the heterodimeric DNA damage-binding (DDB) complex while another protein (p48) forms the small subunit. This protein complex functions in nucleotide-excision repair and binds to DNA following UV damage. Defective activity of this complex causes the repair defect in patients with xeroderma pigmentosum complementation group E (XPE) - an autosomal recessive disorder characterized by photosensitivity and early onset of carcinomas. However, it remains for mutation analysis to demonstrate whether the defect in XPE patients is in this gene or the gene encoding the small subunit. In addition, Best vitelliform mascular dystrophy is mapped to the same region as this gene on 11q, but no sequence alternations of this gene are demonstrated in Best disease patients. The protein encoded by this gene also functions as an adaptor molecule for the cullin 4 (CUL4) ubiquitin E3 ligase complex by facilitating the binding of substrates to this complex and the ubiquitination of proteins. [provided by RefSeq, May 2012] | ||||
RNA binding domains (RBDs)
| Protein | Domain | Pfam ID | E-value | Domain number |
|---|---|---|---|---|
| ENSP00000506457 | CPSF_A | PF03178 | 4e-86 | 1 |
| ENSP00000504954 | CPSF_A | PF03178 | 4.5e-86 | 1 |
| ENSP00000506134 | CPSF_A | PF03178 | 4.7e-86 | 1 |
| ENSP00000504878 | CPSF_A | PF03178 | 5.2e-86 | 1 |
| ENSP00000301764 | CPSF_A | PF03178 | 5.4e-86 | 1 |
| ENSP00000506223 | CPSF_A | PF03178 | 5.4e-86 | 1 |
| ENSP00000506685 | CPSF_A | PF03178 | 5.4e-86 | 1 |
| ENSP00000505437 | CPSF_A | PF03178 | 6e-86 | 1 |
| ENSP00000440269 | CPSF_A | PF03178 | 3.1e-77 | 1 |
| ENSP00000505717 | CPSF_A | PF03178 | 6.8e-73 | 1 |
| ENSP00000445844 | CPSF_A | PF03178 | 1.2e-09 | 1 |
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|---|---|---|---|
| 36333315 | Translation regulatory factor BZW1 regulates preimplantation embryo development and compaction by restricting global non-AUG Initiation. | Jue Zhang | 2022-11-04 | Nature communications |
| 22319459 | Raf1 Is a DCAF for the Rik1 DDB1-like protein and has separable roles in siRNA generation and chromatin modification. | Alessia Buscaino | 2012-02-01 | PLoS genetics |
| 30704981 | Inhibition of HBV Transcription From cccDNA With Nitazoxanide by Targeting the HBx-DDB1 Interaction. | Kazuma Sekiba | 2019-01-01 | Cellular and molecular gastroenterology and hepatology |
| 24206017 | Measuring cereblon as a biomarker of response or resistance to lenalidomide and pomalidomide requires use of standardized reagents and understanding of gene complexity. | K Anita Gandhi | 2014-01-01 | British journal of haematology |
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| DDB1-201 | ENST00000301764 | 4245 | 1140aa | ENSP00000301764 |
| DDB1-225 | ENST00000679588 | 4184 | 1084aa | ENSP00000506134 |
| DDB1-228 | ENST00000680075 | 3981 | 1066aa | ENSP00000504954 |
| DDB1-226 | ENST00000679712 | 4033 | 1069aa | ENSP00000504878 |
| DDB1-232 | ENST00000680357 | 4363 | 217aa | ENSP00000506385 |
| DDB1-236 | ENST00000680717 | 3937 | 1027aa | ENSP00000506457 |
| DDB1-233 | ENST00000680367 | 4426 | 1140aa | ENSP00000506223 |
| DDB1-243 | ENST00000681803 | 4348 | 1140aa | ENSP00000506685 |
| DDB1-234 | ENST00000680452 | 3110 | No protein | - |
| DDB1-206 | ENST00000535283 | 4920 | 370aa | ENSP00000441825 |
| DDB1-240 | ENST00000681188 | 4329 | 78aa | ENSP00000506472 |
| DDB1-235 | ENST00000680602 | 4001 | 1061aa | ENSP00000505437 |
| DDB1-237 | ENST00000680881 | 4335 | No protein | - |
| DDB1-242 | ENST00000681580 | 1629 | No protein | - |
| DDB1-244 | ENST00000681815 | 2205 | No protein | - |
| DDB1-238 | ENST00000680959 | 3114 | No protein | - |
| DDB1-245 | ENST00000681935 | 4138 | 1107aa | ENSP00000505717 |
| DDB1-230 | ENST00000680250 | 4459 | No protein | - |
| DDB1-231 | ENST00000680345 | 2623 | No protein | - |
| DDB1-203 | ENST00000451943 | 1224 | No protein | - |
| DDB1-223 | ENST00000545894 | 2282 | No protein | - |
| DDB1-217 | ENST00000540166 | 3906 | 1092aa | ENSP00000440269 |
| DDB1-212 | ENST00000538470 | 858 | No protein | - |
| DDB1-227 | ENST00000679855 | 3902 | No protein | - |
| DDB1-213 | ENST00000539332 | 1767 | No protein | - |
| DDB1-241 | ENST00000681368 | 3088 | No protein | - |
| DDB1-222 | ENST00000543658 | 571 | 162aa | ENSP00000445844 |
| DDB1-204 | ENST00000535147 | 2789 | No protein | - |
| DDB1-209 | ENST00000537877 | 1375 | No protein | - |
| DDB1-202 | ENST00000414411 | 1904 | No protein | - |
| DDB1-207 | ENST00000535967 | 1404 | No protein | - |
| DDB1-216 | ENST00000539739 | 1624 | No protein | - |
| DDB1-218 | ENST00000540784 | 1119 | No protein | - |
| DDB1-211 | ENST00000538280 | 227 | No protein | - |
| DDB1-224 | ENST00000545930 | 1278 | No protein | - |
| DDB1-239 | ENST00000681067 | 4180 | No protein | - |
| DDB1-229 | ENST00000680096 | 2873 | No protein | - |
| DDB1-205 | ENST00000535174 | 896 | No protein | - |
| DDB1-215 | ENST00000539712 | 1020 | No protein | - |
| DDB1-219 | ENST00000541513 | 958 | No protein | - |
| DDB1-221 | ENST00000543162 | 2592 | No protein | - |
| DDB1-208 | ENST00000537120 | 1215 | No protein | - |
| DDB1-214 | ENST00000539426 | 805 | 165aa | ENSP00000445554 |
| DDB1-210 | ENST00000538129 | 475 | No protein | - |
| DDB1-220 | ENST00000542337 | 558 | 146aa | ENSP00000444105 |
Phenotypes
| ensgID | Trait | pValue | Pubmed ID |
|---|
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
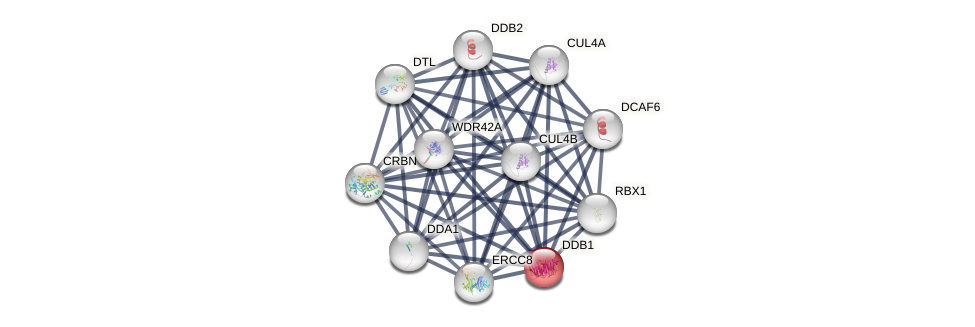
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000167986 | homo_sapiens | ENSP00000305790 | 41.9885 | 22.7609 | homo_sapiens | ENSP00000301764 | 44.8246 | 24.2982 |
| ENSG00000167986 | homo_sapiens | ENSP00000484669 | 32.6403 | 16.7013 | homo_sapiens | ENSP00000301764 | 41.3158 | 21.1404 |
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 98.1579 | 97.8947 | nomascus_leucogenys | ENSNLEP00000002855 | 98.2441 | 97.9807 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.1228 | 99.0351 | pan_paniscus | ENSPPAP00000024544 | 98.5179 | 98.4307 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 100 | 100 | pongo_abelii | ENSPPYP00000003683 | 100 | 100 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.1228 | 99.0351 | pan_troglodytes | ENSPTRP00000081878 | 98.5179 | 98.4307 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.1228 | 99.0351 | gorilla_gorilla | ENSGGOP00000048367 | 98.5179 | 98.4307 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 98.7719 | 97.9825 | ornithorhynchus_anatinus | ENSOANP00000007759 | 98.7719 | 97.9825 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.2105 | 99.0351 | sarcophilus_harrisii | ENSSHAP00000013570 | 97.0815 | 96.9099 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 88.7719 | 88.4211 | notamacropus_eugenii | ENSMEUP00000001016 | 90.4379 | 90.0804 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 76.4912 | 76.2281 | choloepus_hoffmanni | ENSCHOP00000010080 | 79.4895 | 79.216 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 94.1228 | 93.6842 | dasypus_novemcinctus | ENSDNOP00000011548 | 97.192 | 96.7391 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 76.5789 | 76.3158 | erinaceus_europaeus | ENSEEUP00000002953 | 76.7135 | 76.4499 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 61.9298 | 61.5789 | echinops_telfairi | ENSETEP00000000855 | 61.9842 | 61.633 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 100 | 100 | callithrix_jacchus | ENSCJAP00000035865 | 100 | 100 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 98.6842 | 98.5088 | cercocebus_atys | ENSCATP00000044056 | 98.4252 | 98.2502 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.8246 | 99.8246 | macaca_fascicularis | ENSMFAP00000030087 | 100 | 100 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.0351 | 98.9474 | macaca_mulatta | ENSMMUP00000069687 | 98.4307 | 98.3435 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.1228 | 99.0351 | macaca_nemestrina | ENSMNEP00000015252 | 98.5179 | 98.4307 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 100 | 100 | papio_anubis | ENSPANP00000000430 | 100 | 100 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.1228 | 99.0351 | mandrillus_leucophaeus | ENSMLEP00000040250 | 98.5179 | 98.4307 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 85.7895 | 85.3509 | tursiops_truncatus | ENSTTRP00000010491 | 87.4776 | 87.0304 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.9123 | 99.8246 | vulpes_vulpes | ENSVVUP00000006429 | 99.9123 | 99.8246 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.8246 | 99.7368 | mus_spretus | MGP_SPRETEiJ_P0050121 | 99.8246 | 99.7368 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.7368 | 99.7368 | equus_asinus | ENSEASP00005011644 | 99.7368 | 99.7368 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.8246 | 99.7368 | capra_hircus | ENSCHIP00000013716 | 99.8246 | 99.7368 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.8246 | 99.6491 | loxodonta_africana | ENSLAFP00000009624 | 99.8246 | 99.6491 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.9123 | 99.9123 | panthera_leo | ENSPLOP00000008688 | 99.9123 | 99.9123 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.9123 | 99.9123 | ailuropoda_melanoleuca | ENSAMEP00000017485 | 95.4736 | 95.4736 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.2982 | 99.1228 | bos_indicus_hybrid | ENSBIXP00005003242 | 79.7183 | 79.5775 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.9123 | 99.9123 | oryctolagus_cuniculus | ENSOCUP00000001429 | 99.9123 | 99.9123 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.6491 | 99.6491 | equus_caballus | ENSECAP00000074581 | 88.3359 | 88.3359 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.9123 | 99.9123 | canis_lupus_familiaris | ENSCAFP00845018193 | 99.9123 | 99.9123 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.9123 | 99.9123 | panthera_pardus | ENSPPRP00000022952 | 99.9123 | 99.9123 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.9123 | 99.8246 | marmota_marmota_marmota | ENSMMMP00000001387 | 99.9123 | 99.8246 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.8246 | 99.4737 | balaenoptera_musculus | ENSBMSP00010010513 | 95.3099 | 94.9749 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.6491 | 99.1228 | cavia_porcellus | ENSCPOP00000027409 | 99.6491 | 99.1228 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.9123 | 99.9123 | felis_catus | ENSFCAP00000024813 | 99.9123 | 99.9123 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 94.1228 | 94.0351 | ursus_americanus | ENSUAMP00000022657 | 93.2233 | 93.1364 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 89.1228 | 88.5965 | monodelphis_domestica | ENSMODP00000025457 | 99.0253 | 98.4405 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.8246 | 99.7368 | bison_bison_bison | ENSBBBP00000024789 | 99.8246 | 99.7368 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.8246 | 99.4737 | monodon_monoceros | ENSMMNP00015023883 | 99.8246 | 99.4737 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.7368 | 99.6491 | sus_scrofa | ENSSSCP00000013928 | 99.8244 | 99.7366 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.9123 | 99.8246 | microcebus_murinus | ENSMICP00000012517 | 99.9123 | 99.8246 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.4737 | 98.7719 | octodon_degus | ENSODEP00000003153 | 99.2995 | 98.5989 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.8246 | 99.6491 | jaculus_jaculus | ENSJJAP00000015706 | 99.8246 | 99.6491 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.4737 | 99.1228 | ovis_aries_rambouillet | ENSOARP00020015969 | 91.5254 | 91.2026 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 94.1228 | 94.0351 | ursus_maritimus | ENSUMAP00000016040 | 97.7231 | 97.6321 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 84.5614 | 84.0351 | ochotona_princeps | ENSOPRP00000007573 | 84.5614 | 84.0351 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 98.4211 | 97.807 | delphinapterus_leucas | ENSDLEP00000001400 | 95.8155 | 95.2178 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.8246 | 99.4737 | physeter_catodon | ENSPCTP00005028154 | 99.8246 | 99.4737 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.9123 | 99.7368 | mus_caroli | MGP_CAROLIEiJ_P0048141 | 99.9123 | 99.7368 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.7368 | 99.6491 | mus_musculus | ENSMUSP00000025649 | 99.7368 | 99.6491 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 84.1228 | 82.0175 | dipodomys_ordii | ENSDORP00000000116 | 93.561 | 91.2195 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 96.6667 | 96.5789 | mustela_putorius_furo | ENSMPUP00000014091 | 98.9228 | 98.833 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 100 | 100 | aotus_nancymaae | ENSANAP00000007973 | 100 | 100 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.9123 | 99.9123 | saimiri_boliviensis_boliviensis | ENSSBOP00000006064 | 99.9123 | 99.9123 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 86.0526 | 85.9649 | vicugna_pacos | ENSVPAP00000007407 | 87.9032 | 87.8136 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 100 | 100 | chlorocebus_sabaeus | ENSCSAP00000002685 | 100 | 100 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.9123 | 99.8246 | mesocricetus_auratus | ENSMAUP00000012727 | 99.9123 | 99.8246 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.8246 | 99.386 | phocoena_sinus | ENSPSNP00000017573 | 99.8246 | 99.386 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.8246 | 99.7368 | camelus_dromedarius | ENSCDRP00005013126 | 99.8246 | 99.7368 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.4737 | 99.1228 | carlito_syrichta | ENSTSYP00000007462 | 99.6485 | 99.297 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 98.0702 | 98.0702 | panthera_tigris_altaica | ENSPTIP00000004430 | 98.0702 | 98.0702 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.9123 | 99.7368 | rattus_norvegicus | ENSRNOP00000028188 | 99.9123 | 99.7368 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.9123 | 99.7368 | prolemur_simus | ENSPSMP00000030622 | 99.9123 | 99.7368 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.9123 | 99.5614 | microtus_ochrogaster | ENSMOCP00000023924 | 99.9123 | 99.5614 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 63.6842 | 63.0702 | sorex_araneus | ENSSARP00000003012 | 66.9124 | 66.2673 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.9123 | 99.8246 | peromyscus_maniculatus_bairdii | ENSPEMP00000011695 | 99.9123 | 99.8246 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.8246 | 99.7368 | bos_mutus | ENSBMUP00000022875 | 99.8246 | 99.7368 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.8246 | 99.7368 | mus_spicilegus | ENSMSIP00000036624 | 98.7847 | 98.6979 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.9123 | 99.8246 | cricetulus_griseus_chok1gshd | ENSCGRP00001016252 | 99.9123 | 99.8246 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.8246 | 99.7368 | pteropus_vampyrus | ENSPVAP00000003641 | 99.8246 | 99.7368 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.5614 | 99.2982 | myotis_lucifugus | ENSMLUP00000012054 | 99.6488 | 99.3854 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.4737 | 99.2105 | vombatus_ursinus | ENSVURP00010004154 | 99.4737 | 99.2105 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.8246 | 99.7368 | rhinolophus_ferrumequinum | ENSRFEP00010002421 | 99.8246 | 99.7368 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.9123 | 99.9123 | neovison_vison | ENSNVIP00000028189 | 99.9123 | 99.9123 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.8246 | 99.7368 | bos_taurus | ENSBTAP00000028740 | 93.0499 | 92.9681 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 39.386 | 39.2982 | heterocephalus_glaber_female | ENSHGLP00000025896 | 99.7778 | 99.5556 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 100 | 100 | rhinopithecus_bieti | ENSRBIP00000019339 | 100 | 100 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 94.1228 | 94.0351 | canis_lupus_dingo | ENSCAFP00020015849 | 98.9852 | 98.893 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.9123 | 99.7368 | sciurus_vulgaris | ENSSVLP00005011413 | 99.9123 | 99.7368 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.4737 | 99.0351 | phascolarctos_cinereus | ENSPCIP00000003212 | 99.4737 | 99.0351 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 81.0526 | 80.614 | procavia_capensis | ENSPCAP00000010463 | 81.5534 | 81.1121 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.6491 | 99.386 | nannospalax_galili | ENSNGAP00000006337 | 99.6491 | 99.386 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.7368 | 99.2105 | chinchilla_lanigera | ENSCLAP00000005820 | 99.7368 | 99.2105 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.2982 | 99.1228 | cervus_hanglu_yarkandensis | ENSCHYP00000014313 | 98.2639 | 98.0903 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.6491 | 99.386 | catagonus_wagneri | ENSCWAP00000029063 | 99.6491 | 99.386 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.9123 | 99.8246 | ictidomys_tridecemlineatus | ENSSTOP00000006801 | 99.9123 | 99.8246 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.8246 | 99.6491 | otolemur_garnettii | ENSOGAP00000014421 | 99.8246 | 99.6491 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 100 | 100 | cebus_imitator | ENSCCAP00000036063 | 100 | 100 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.8246 | 99.7368 | urocitellus_parryii | ENSUPAP00010005335 | 99.8246 | 99.7368 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.8246 | 99.6491 | moschus_moschiferus | ENSMMSP00000019907 | 99.8246 | 99.6491 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.1228 | 98.9474 | rhinopithecus_roxellana | ENSRROP00000036042 | 98.5179 | 98.3435 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 99.9123 | 99.7368 | mus_pahari | MGP_PahariEiJ_P0021136 | 99.9123 | 99.7368 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 98.7719 | 98.5088 | propithecus_coquereli | ENSPCOP00000007867 | 98.5989 | 98.3363 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 61.6667 | 38.2456 | caenorhabditis_elegans | M18.5.1 | 61.9929 | 38.448 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 76.4035 | 61.0526 | drosophila_melanogaster | FBpp0082177 | 76.4035 | 61.0526 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 77.7193 | 62.5439 | ciona_intestinalis | ENSCINP00000015292 | 77.3799 | 62.2707 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 84.0351 | 77.4561 | petromyzon_marinus | ENSPMAP00000009366 | 94.1061 | 86.7387 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 72.7193 | 67.1053 | eptatretus_burgeri | ENSEBUP00000023727 | 91.7035 | 84.6239 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 95.8772 | 92.193 | callorhinchus_milii | ENSCMIP00000026284 | 95.542 | 91.8706 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 96.8421 | 93.6842 | latimeria_chalumnae | ENSLACP00000020033 | 96.6725 | 93.5201 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 81.9298 | 76.0526 | lepisosteus_oculatus | ENSLOCP00000000018 | 89.4636 | 83.046 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 95.5263 | 91.1404 | clupea_harengus | ENSCHAP00000012285 | 95.5263 | 91.1404 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 93.5965 | 88.3333 | danio_rerio | ENSDARP00000130126 | 93.4326 | 88.1786 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 13.4211 | 12.3684 | carassius_auratus | ENSCARP00000088616 | 91.0714 | 83.9286 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 95.7895 | 91.3158 | carassius_auratus | ENSCARP00000050740 | 95.7895 | 91.3158 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 25.614 | 24.2982 | astyanax_mexicanus | ENSAMXP00000052495 | 96.0526 | 91.1184 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 95.7018 | 90.1754 | ictalurus_punctatus | ENSIPUP00000008267 | 95.7018 | 90.1754 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 95.3509 | 90.7895 | esox_lucius | ENSELUP00000025124 | 95.4346 | 90.8692 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 94.9123 | 89.7368 | electrophorus_electricus | ENSEEEP00000002205 | 94.333 | 89.1892 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 95 | 90.5263 | oncorhynchus_kisutch | ENSOKIP00005012475 | 94.9167 | 90.447 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 94.1228 | 89.2982 | oncorhynchus_kisutch | ENSOKIP00005045252 | 93.958 | 89.1419 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 95 | 90.5263 | oncorhynchus_mykiss | ENSOMYP00000029392 | 94.9167 | 90.447 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 94.4737 | 89.386 | oncorhynchus_mykiss | ENSOMYP00000055477 | 94.2257 | 89.1514 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 94.2982 | 89.9123 | salmo_salar | ENSSSAP00000098959 | 93.8045 | 89.4415 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 95.4386 | 90.5263 | salmo_salar | ENSSSAP00000076675 | 95.5224 | 90.6058 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 94.2982 | 89.9123 | salmo_trutta | ENSSTUP00000027596 | 93.8045 | 89.4415 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 95.3509 | 90.614 | salmo_trutta | ENSSTUP00000010396 | 95.4346 | 90.6936 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 95 | 89.8246 | gadus_morhua | ENSGMOP00000020854 | 95.2507 | 90.0616 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 85.1754 | 81.1404 | fundulus_heteroclitus | ENSFHEP00000034388 | 94.2718 | 89.8058 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 91.0526 | 86.4035 | poecilia_reticulata | ENSPREP00000006475 | 94.1924 | 89.3829 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 90.5263 | 86.0526 | xiphophorus_maculatus | ENSXMAP00000000022 | 95.4672 | 90.7493 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 92.2807 | 86.4035 | oryzias_latipes | ENSORLP00000001737 | 93.4281 | 87.4778 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 95.7895 | 90.8772 | cyclopterus_lumpus | ENSCLMP00005001621 | 95.6217 | 90.718 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 95.7895 | 91.4035 | oreochromis_niloticus | ENSONIP00000071715 | 95.7895 | 91.4035 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 95.7895 | 91.4035 | haplochromis_burtoni | ENSHBUP00000007296 | 95.7895 | 91.4035 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 95.7895 | 91.4035 | astatotilapia_calliptera | ENSACLP00000018313 | 95.7895 | 91.4035 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 96.0526 | 91.4912 | sparus_aurata | ENSSAUP00010004485 | 96.0526 | 91.4912 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 95 | 90 | lates_calcarifer | ENSLCAP00010040539 | 95.2507 | 90.2375 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 95.5263 | 92.3684 | xenopus_tropicalis | ENSXETP00000109123 | 94.5312 | 91.4062 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 98.9474 | 97.6316 | chrysemys_picta_bellii | ENSCPBP00000024940 | 98.9474 | 97.6316 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 98.8596 | 97.807 | sphenodon_punctatus | ENSSPUP00000025248 | 98.9464 | 97.8929 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 94.2982 | 91.6667 | laticauda_laticaudata | ENSLLTP00000002619 | 91.6454 | 89.0878 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 98.5088 | 96.7544 | notechis_scutatus | ENSNSUP00000004345 | 98.5088 | 96.7544 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 98.5088 | 96.7544 | pseudonaja_textilis | ENSPTXP00000018372 | 98.5088 | 96.7544 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 89.5614 | 88.0702 | anas_platyrhynchos_platyrhynchos | ENSAPLP00000006857 | 97.2381 | 95.619 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 93.5965 | 91.4912 | gallus_gallus | ENSGALP00010028369 | 95.6951 | 93.5426 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 92.2807 | 89.9123 | meleagris_gallopavo | ENSMGAP00000004938 | 91.8777 | 89.5197 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 94.1228 | 92.3684 | serinus_canaria | ENSSCAP00000013199 | 96.5797 | 94.7795 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 94.0351 | 92.3684 | parus_major | ENSPMJP00000008394 | 96.4896 | 94.7795 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 98.5088 | 96.9298 | anolis_carolinensis | ENSACAP00000017490 | 98.5088 | 96.9298 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 95.8772 | 91.4912 | neolamprologus_brichardi | ENSNBRP00000025658 | 95.8772 | 91.4912 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 98.5088 | 96.8421 | naja_naja | ENSNNAP00000000528 | 98.5088 | 96.8421 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 98.4211 | 96.6667 | podarcis_muralis | ENSPMRP00000002701 | 98.4211 | 96.6667 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 89.2982 | 87.7193 | geospiza_fortis | ENSGFOP00000016578 | 96.4929 | 94.7867 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 98.8596 | 97.6316 | taeniopygia_guttata | ENSTGUP00000006648 | 98.6865 | 97.4606 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 87.2807 | 83.2456 | erpetoichthys_calabaricus | ENSECRP00000030483 | 93.6911 | 89.3597 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 90.4386 | 86.0526 | kryptolebias_marmoratus | ENSKMAP00000003119 | 95.907 | 91.2558 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 98.5965 | 97.1053 | salvator_merianae | ENSSMRP00000002693 | 98.5965 | 97.1053 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 95.4386 | 90.3509 | oryzias_javanicus | ENSOJAP00000001275 | 95.859 | 90.7489 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 82.5439 | 78.8596 | amphilophus_citrinellus | ENSACIP00000026665 | 95.9225 | 91.6412 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 96.0526 | 91.4912 | dicentrarchus_labrax | ENSDLAP00005051979 | 96.0526 | 91.4912 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 96.0526 | 91.1404 | amphiprion_percula | ENSAPEP00000000123 | 96.0526 | 91.1404 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 93.4211 | 88.2456 | oryzias_sinensis | ENSOSIP00000007981 | 93.9982 | 88.7908 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 97.1053 | 95.614 | pelodiscus_sinensis | ENSPSIP00000015016 | 98.4875 | 96.9751 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 98.9474 | 97.7193 | gopherus_evgoodei | ENSGEVP00005009010 | 98.9474 | 97.7193 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 97.193 | 95.5263 | chelonoidis_abingdonii | ENSCABP00000012302 | 97.7072 | 96.0317 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 96.0526 | 91.3158 | seriola_dumerili | ENSSDUP00000006352 | 96.0526 | 91.3158 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 96.0526 | 91.3158 | cottoperca_gobio | ENSCGOP00000006481 | 96.0526 | 91.3158 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 97.2807 | 95.7895 | struthio_camelus_australis | ENSSCUP00000000150 | 98.0548 | 96.5517 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 93.2456 | 91.9298 | anser_brachyrhynchus | ENSABRP00000026177 | 98.6085 | 97.2171 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 91.0526 | 86.6667 | gasterosteus_aculeatus | ENSGACP00000022712 | 95.1421 | 90.5591 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 91.2281 | 85.7895 | cynoglossus_semilaevis | ENSCSEP00000019584 | 91.4688 | 86.0158 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 95.4386 | 90.7895 | poecilia_formosa | ENSPFOP00000003124 | 95.3549 | 90.7099 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 94.4737 | 89.386 | myripristis_murdjan | ENSMMDP00005050064 | 93.6522 | 88.6087 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 95.9649 | 91.4035 | sander_lucioperca | ENSSLUP00000038401 | 95.9649 | 91.4035 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 98.7719 | 97.4561 | strigops_habroptila | ENSSHBP00005022189 | 98.7719 | 97.4561 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 95.8772 | 91.0526 | amphiprion_ocellaris | ENSAOCP00000019570 | 95.8772 | 91.0526 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 98.9474 | 97.7193 | terrapene_carolina_triunguis | ENSTMTP00000020574 | 98.9474 | 97.7193 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 95.3509 | 90.3509 | hucho_hucho | ENSHHUP00000041610 | 95.1006 | 90.1137 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 95.1754 | 90.614 | hucho_hucho | ENSHHUP00000035553 | 95.092 | 90.5346 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 96.0526 | 91.0526 | betta_splendens | ENSBSLP00000025222 | 96.0526 | 91.0526 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 91.5789 | 86.3158 | pygocentrus_nattereri | ENSPNAP00000037661 | 94.3942 | 88.9693 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 94.2982 | 89.5614 | anabas_testudineus | ENSATEP00000029888 | 94.7137 | 89.9559 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 72.1053 | 57.1053 | ciona_savignyi | ENSCSAVP00000008027 | 76.1816 | 60.3336 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 25 | 12.5439 | saccharomyces_cerevisiae | YML049C | 20.9405 | 10.507 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 31.0526 | 13.0702 | saccharomyces_cerevisiae | YDR301W | 26.087 | 10.9801 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 96.0526 | 91.3158 | seriola_lalandi_dorsalis | ENSSLDP00000020241 | 96.0526 | 91.3158 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 81.4035 | 77.4561 | labrus_bergylta | ENSLBEP00000022242 | 95.5716 | 90.9372 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 46.9298 | 44.7368 | pundamilia_nyererei | ENSPNYP00000032109 | 93.3682 | 89.0052 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 47.0175 | 44.386 | pundamilia_nyererei | ENSPNYP00000013871 | 95.3737 | 90.0356 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 96.0526 | 91.4035 | mastacembelus_armatus | ENSMAMP00000064966 | 96.0526 | 91.4035 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 91.0526 | 86.4912 | stegastes_partitus | ENSSPAP00000013106 | 95.6682 | 90.8756 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 94.5614 | 90.0877 | oncorhynchus_tshawytscha | ENSOTSP00005055139 | 94.1485 | 89.6943 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 93.1579 | 88.0702 | oncorhynchus_tshawytscha | ENSOTSP00005048613 | 93.6508 | 88.5362 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 93.1579 | 88.0702 | oncorhynchus_tshawytscha | ENSOTSP00005100505 | 93.6508 | 88.5362 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 94.4737 | 89.4737 | acanthochromis_polyacanthus | ENSAPOP00000023266 | 93.5708 | 88.6186 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 94.2982 | 92.193 | aquila_chrysaetos_chrysaetos | ENSACCP00020017798 | 93.5596 | 91.4708 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 95.5263 | 90.8772 | nothobranchius_furzeri | ENSNFUP00015024546 | 95.1092 | 90.4803 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 90.1754 | 86.1404 | cyprinodon_variegatus | ENSCVAP00000016510 | 95.1852 | 90.9259 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 95.5263 | 90.7018 | denticeps_clupeoides | ENSDCDP00000058340 | 95.1092 | 90.3057 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 95.2632 | 90.614 | larimichthys_crocea | ENSLCRP00005036697 | 95.2632 | 90.614 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 93.1579 | 87.193 | takifugu_rubripes | ENSTRUP00000042679 | 93.1579 | 87.193 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 86.3158 | 84.6491 | ficedula_albicollis | ENSFALP00000012512 | 94.1627 | 92.3445 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 90.7895 | 86.0526 | hippocampus_comes | ENSHCOP00000015095 | 94.6935 | 89.753 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 95.614 | 90.7018 | cyprinus_carpio_carpio | ENSCCRP00000001393 | 95.614 | 90.7018 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 91.0526 | 86.4912 | cyprinus_carpio_carpio | ENSCCRP00000001428 | 94.6217 | 89.8815 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 92.807 | 91.2281 | coturnix_japonica | ENSCJPP00005015983 | 97.1534 | 95.5005 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 95.3509 | 90.614 | poecilia_latipinna | ENSPLAP00000013643 | 95.4346 | 90.6936 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 89.2982 | 84.2982 | sinocyclocheilus_grahami | ENSSGRP00000041925 | 91.137 | 86.034 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 95.7895 | 91.3158 | maylandia_zebra | ENSMZEP00005018434 | 95.7895 | 91.3158 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 77.3684 | 71.2281 | tetraodon_nigroviridis | ENSTNIP00000012438 | 88.0239 | 81.0379 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 91.7544 | 86.8421 | paramormyrops_kingsleyae | ENSPKIP00000000663 | 91.5937 | 86.69 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 95.4386 | 90.7018 | scleropages_formosus | ENSSFOP00015017367 | 95.6063 | 90.8612 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 96.4912 | 92.8947 | leptobrachium_leishanense | ENSLLEP00000032402 | 96.4912 | 92.8947 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 94.6491 | 89.4737 | scophthalmus_maximus | ENSSMAP00000015534 | 93.8261 | 88.6957 |
| ENSG00000167986 | homo_sapiens | ENSP00000301764 | 91.7544 | 86.5789 | oryzias_melastigma | ENSOMEP00000005155 | 94.6606 | 89.3213 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0000781 | located_in chromosome, telomeric region | 1 | HDA | Homo_sapiens(9606) | Component |
| GO:0003677 | enables DNA binding | 1 | TAS | Homo_sapiens(9606) | Function |
| GO:0003684 | contributes_to damaged DNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005615 | located_in extracellular space | 1 | HDA | Homo_sapiens(9606) | Component |
| GO:0005634 | is_active_in nucleus | 4 | IBA,IDA,NAS | Homo_sapiens(9606) | Component |
| GO:0005654 | located_in nucleoplasm | 2 | IDA,TAS | Homo_sapiens(9606) | Component |
| GO:0005730 | is_active_in nucleolus | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005737 | located_in cytoplasm | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0006281 | involved_in DNA repair | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0006289 | involved_in nucleotide-excision repair | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0006511 | involved_in ubiquitin-dependent protein catabolic process | 2 | IDA,IMP | Homo_sapiens(9606) | Process |
| GO:0006915 | involved_in apoptotic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0006974 | involved_in DNA damage response | 3 | EXP,IMP,ISS | Homo_sapiens(9606) | Process |
| GO:0007056 | involved_in spindle assembly involved in female meiosis | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0010498 | acts_upstream_of_or_within proteasomal protein catabolic process | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0016055 | involved_in Wnt signaling pathway | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0016567 | involved_in protein ubiquitination | 3 | IDA,IEA,IMP | Homo_sapiens(9606) | Process |
| GO:0019076 | involved_in viral release from host cell | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0030674 | enables protein-macromolecule adaptor activity | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0031464 | part_of Cul4A-RING E3 ubiquitin ligase complex | 2 | EXP,IDA | Homo_sapiens(9606) | Component |
| GO:0031465 | part_of Cul4B-RING E3 ubiquitin ligase complex | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0032991 | part_of protein-containing complex | 1 | IMP | Homo_sapiens(9606) | Component |
| GO:0034644 | involved_in cellular response to UV | 2 | EXP,ISS | Homo_sapiens(9606) | Process |
| GO:0035234 | involved_in ectopic germ cell programmed cell death | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0035861 | is_active_in site of double-strand break | 1 | IBA | Homo_sapiens(9606) | Component |
| GO:0042752 | involved_in regulation of circadian rhythm | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0043066 | involved_in negative regulation of apoptotic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0043161 | involved_in proteasome-mediated ubiquitin-dependent protein catabolic process | 3 | IBA,IDA,IMP | Homo_sapiens(9606) | Process |
| GO:0044725 | involved_in epigenetic programming in the zygotic pronuclei | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0044877 | enables protein-containing complex binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0045070 | involved_in positive regulation of viral genome replication | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0045722 | involved_in positive regulation of gluconeogenesis | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0045732 | involved_in positive regulation of protein catabolic process | 2 | IDA,IMP | Homo_sapiens(9606) | Process |
| GO:0046726 | involved_in positive regulation by virus of viral protein levels in host cell | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0048511 | involved_in rhythmic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0051093 | involved_in negative regulation of developmental process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0051702 | involved_in biological process involved in interaction with symbiont | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0070062 | located_in extracellular exosome | 1 | HDA | Homo_sapiens(9606) | Component |
| GO:0070914 | involved_in UV-damage excision repair | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0071987 | enables WD40-repeat domain binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0080008 | part_of Cul4-RING E3 ubiquitin ligase complex | 2 | IDA,IMP | Homo_sapiens(9606) | Component |
| GO:0097602 | enables cullin family protein binding | 2 | IDA,IPI | Homo_sapiens(9606) | Function |
| GO:0160072 | enables ubiquitin ligase complex scaffold activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:1901990 | involved_in regulation of mitotic cell cycle phase transition | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:2000242 | involved_in negative regulation of reproductive process | 1 | IEA | Homo_sapiens(9606) | Process |