Welcome to
RBP World!
Functions and Diseases
| RBP Type | Canonical_RBPs |
| Diseases | CancerSARS-COV-OC43FlavivirusesHIVRVSARS-COV-2SINVVEEVDengueZika |
| Drug | N.A. |
| Main interacting RNAs | mRNA |
| Moonlighting functions | N.A. |
| Localizations | Stress granucle |
| BulkPerturb-seq | N.A. |
Description
| Ensembl ID | ENSG00000162613 | Gene ID | 8880 | Accession | 4004 |
| Symbol | FUBP1 | Alias | FBP;FUBP;hDHV | Full Name | far upstream element binding protein 1 |
| Status | Confidence | Length | 35056 bases | Strand | Minus strand |
| Position | 1 : 77944055 - 77979110 | RNA binding domain | KH_2 , DUF1897 , KH_1 | ||
| Summary | The protein encoded by this gene is a single stranded DNA-binding protein that binds to multiple DNA elements, including the far upstream element (FUSE) located upstream of c-myc. Binding to FUSE occurs on the non-coding strand, and is important to the regulation of c-myc in undifferentiated cells. This protein contains three domains, an amphipathic helix N-terminal domain, a DNA-binding central domain, and a C-terminal transactivation domain that contains three tyrosine-rich motifs. The N-terminal domain is thought to repress the activity of the C-terminal domain. This protein is also thought to bind RNA, and contains 3'-5' helicase activity with in vitro activity on both DNA-DNA and RNA-RNA duplexes. Aberrant expression of this gene has been found in malignant tissues, and this gene is important to neural system and lung development. Binding of this protein to viral RNA is thought to play a role in several viral diseases, including hepatitis C and hand, foot and mouth disease. Alternative splicing results in multiple transcript variants. [provided by RefSeq, Dec 2014] | ||||
RNA binding domains (RBDs)
| Protein | Domain | Pfam ID | E-value | Domain number |
|---|---|---|---|---|
| ENSP00000359804 | KH_2 | PF07650 | 4e-22 | 1 |
| ENSP00000359804 | KH_2 | PF07650 | 4e-22 | 2 |
| ENSP00000359804 | KH_1 | PF00013 | 1.2e-68 | 3 |
| ENSP00000359804 | KH_1 | PF00013 | 1.2e-68 | 4 |
| ENSP00000359804 | KH_1 | PF00013 | 1.2e-68 | 5 |
| ENSP00000359804 | KH_1 | PF00013 | 1.2e-68 | 6 |
| ENSP00000359804 | DUF1897 | PF09005 | 1.8e-10 | 7 |
| ENSP00000359804 | DUF1897 | PF09005 | 1.8e-10 | 8 |
| ENSP00000359803 | KH_2 | PF07650 | 4.4e-22 | 1 |
| ENSP00000359803 | KH_2 | PF07650 | 4.4e-22 | 2 |
| ENSP00000359803 | KH_1 | PF00013 | 1.3e-68 | 3 |
| ENSP00000359803 | KH_1 | PF00013 | 1.3e-68 | 4 |
| ENSP00000359803 | KH_1 | PF00013 | 1.3e-68 | 5 |
| ENSP00000359803 | KH_1 | PF00013 | 1.3e-68 | 6 |
| ENSP00000359803 | DUF1897 | PF09005 | 2e-10 | 7 |
| ENSP00000359803 | DUF1897 | PF09005 | 2e-10 | 8 |
| ENSP00000294623 | KH_2 | PF07650 | 4.7e-22 | 1 |
| ENSP00000294623 | KH_2 | PF07650 | 4.7e-22 | 2 |
| ENSP00000294623 | KH_1 | PF00013 | 1.2e-68 | 3 |
| ENSP00000294623 | KH_1 | PF00013 | 1.2e-68 | 4 |
| ENSP00000294623 | KH_1 | PF00013 | 1.2e-68 | 5 |
| ENSP00000294623 | KH_1 | PF00013 | 1.2e-68 | 6 |
| ENSP00000294623 | DUF1897 | PF09005 | 2e-10 | 7 |
| ENSP00000294623 | DUF1897 | PF09005 | 2e-10 | 8 |
| ENSP00000515388 | KH_2 | PF07650 | 5.2e-22 | 1 |
| ENSP00000515388 | KH_2 | PF07650 | 5.2e-22 | 2 |
| ENSP00000515388 | KH_1 | PF00013 | 1.4e-68 | 3 |
| ENSP00000515388 | KH_1 | PF00013 | 1.4e-68 | 4 |
| ENSP00000515388 | KH_1 | PF00013 | 1.4e-68 | 5 |
| ENSP00000515388 | KH_1 | PF00013 | 1.4e-68 | 6 |
| ENSP00000515388 | DUF1897 | PF09005 | 2.1e-10 | 7 |
| ENSP00000515388 | DUF1897 | PF09005 | 2.1e-10 | 8 |
| ENSP00000402630 | KH_2 | PF07650 | 6.1e-22 | 1 |
| ENSP00000402630 | KH_2 | PF07650 | 6.1e-22 | 2 |
| ENSP00000402630 | KH_1 | PF00013 | 1.6e-68 | 3 |
| ENSP00000402630 | KH_1 | PF00013 | 1.6e-68 | 4 |
| ENSP00000402630 | KH_1 | PF00013 | 1.6e-68 | 5 |
| ENSP00000402630 | KH_1 | PF00013 | 1.6e-68 | 6 |
| ENSP00000402630 | DUF1897 | PF09005 | 2.4e-10 | 7 |
| ENSP00000402630 | DUF1897 | PF09005 | 2.4e-10 | 8 |
| ENSP00000515389 | DUF1897 | PF09005 | 4.8e-10 | 1 |
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|---|---|---|---|
| 36418423 | A mass spectrometry-based approach for the identification of Kpnβ1 binding partners in cancer cells. | O Michael Okpara | 2022-11-23 | Scientific reports |
| 36932454 | circMMD reduction following tumor treating fields inhibits glioblastoma progression through FUBP1/FIR/DVL1 and miR-15b-5p/FZD6 signaling. | Shengchao Xu | 2023-03-17 | Journal of experimental & clinical cancer research : CR |
| 27899653 | Control of the negative IRES trans-acting factor KHSRP by ubiquitination. | Yu-An Kung | 2017-01-09 | Nucleic acids research |
| 33999210 | dSPRINT: predicting DNA, RNA, ion, peptide and small molecule interaction sites within protein domains. | Anat Etzion-Fuchs | 2021-07-21 | Nucleic acids research |
| 29606613 | Far Upstream Element-Binding Protein 1 Regulates LSD1 Alternative Splicing to Promote Terminal Differentiation of Neural Progenitors. | Inah Hwang | 2018-04-10 | Stem cell reports |
| 25662218 | FUBP1: a new protagonist in splicing regulation of the DMD gene. | Julie Miro | 2015-02-27 | Nucleic acids research |
| 35274005 | Pan-Cancer Transcriptome and Immune Infiltration Analyses Reveal the Oncogenic Role of Far Upstream Element-Binding Protein 1 (FUBP1). | Huan Wang | 2022-01-01 | Frontiers in molecular biosciences |
| 32481602 | Multiple Functions of Fubp1 in Cell Cycle Progression and Cell Survival. | Mingyu Kang | 2020-05-28 | Cells |
| 23818605 | Far upstream element-binding protein 1 and RNA secondary structure both mediate second-step splicing repression. | Huang Li | 2013-07-16 | Proceedings of the National Academy of Sciences of the United States of America |
| 34116677 | The circACTN4 interacts with FUBP1 to promote tumorigenesis and progression of breast cancer by regulating the expression of proto-oncogene MYC. | Xiaosong Wang | 2021-06-11 | Molecular cancer |
| 37506698 | FUBP1 is a general splicing factor facilitating 3' splice site recognition and splicing of long introns. | Stefanie Ebersberger | 2023-08-03 | Molecular cell |
| 28937658 | Extracellular Vesicle-Associated RNA as a Carrier of Epigenetic Information. | Maria Carlo CM Di | 2017-09-22 | Genes |
| 26253612 | Genetic Predisposition to Weight Loss and Regain With Lifestyle Intervention: Analyses From the Diabetes Prevention Program and the Look AHEAD Randomized Controlled Trials. | D George Papandonatos | 2015-12-01 | Diabetes |
| 31553912 | Identification of FUBP1 as a Long Tail Cancer Driver and Widespread Regulator of Tumor Suppressor and Oncogene Alternative Splicing. | S Jessica Elman | 2019-09-24 | Cell reports |
| 32555178 | NORAD orchestrates endometrial cancer progression by sequestering FUBP1 nuclear localization to promote cell apoptosis. | Tong Han | 2020-06-18 | Cell death & disease |
| 35558282 | Selective enrichment of sialylated glycopeptides with a d-allose@SiO2 matrix. | Na Sun | 2018-11-16 | RSC advances |
| 34430595 | Identifying a possible new target for diagnosis and treatment of postmenopausal osteoporosis through bioinformatics and clinical sample analysis. | Ting Liu | 2021-07-01 | Annals of translational medicine |
| 30343319 | The master regulator FUBP1: its emerging role in normal cell function and malignant development. | Lydie Debaize | 2019-01-01 | Cellular and molecular life sciences : CMLS |
| 35546072 | Far upstream element -binding protein 1 (FUBP1) participates in the malignant process and glycolysis of colon cancer cells by combining with c-Myc. | Shanwei Wang | 2022-05-01 | Bioengineered |
| 24086756 | Novel CIC point mutations and an exon-spanning, homozygous deletion identified in oligodendroglial tumors by a comprehensive genomic approach including transcriptome sequencing. | Sophie Eisenreich | 2013-01-01 | PloS one |
| 33676361 | Far Upstream Binding Protein 1 (FUBP1) participates in translational regulation of Nrf2 protein under oxidative stress. | Wujing Dai | 2021-05-01 | Redox biology |
| 27157613 | MiR-16 mediates trastuzumab and lapatinib response in ErbB-2-positive breast and gastric cancer via its novel targets CCNJ and FUBP1. | L Venturutti | 2016-12-01 | Oncogene |
| 27566151 | Alternative splicing of U2AF1 reveals a shared repression mechanism for duplicated exons. | Jana Kralovicova | 2017-01-09 | Nucleic acids research |
| 36082445 | A pleiotropic variant in DNAJB4 is associated with multiple myeloma risk. | Marco Dicanio | 2023-01-15 | International journal of cancer |
| 29643205 | In vitro iCLIP-based modeling uncovers how the splicing factor U2AF2 relies on regulation by cofactors. | Reymond X F Sutandy | 2018-05-01 | Genome research |
| 28535583 | Molecular Testing of Brain Tumor. | Sung-Hye Park | 2017-05-01 | Journal of pathology and translational medicine |
| 33185955 | Using protein microarray to identify and evaluate autoantibodies to tumor-associated antigens in ovarian cancer. | Yan Ma | 2021-02-01 | Cancer science |
| 35158909 | RBM22, a Key Player of Pre-mRNA Splicing and Gene Expression Regulation, Is Altered in Cancer. | Benoît Soubise | 2022-01-27 | Cancers |
| 37180822 | Far upstream element-binding protein 1 confers lobaplatin resistance by transcriptionally activating PTGES and facilitating the arachidonic acid metabolic pathway in osteosarcoma. | Qiong Ma | 2023-06-01 | MedComm |
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| FUBP1-208 | ENST00000483894 | 605 | No protein | - |
| FUBP1-207 | ENST00000480673 | 262 | No protein | - |
| FUBP1-206 | ENST00000474632 | 639 | No protein | - |
| FUBP1-211 | ENST00000489495 | 2156 | No protein | - |
| FUBP1-210 | ENST00000488814 | 612 | No protein | - |
| FUBP1-212 | ENST00000492405 | 668 | No protein | - |
| FUBP1-213 | ENST00000492724 | 583 | No protein | - |
| FUBP1-203 | ENST00000370768 | 6714 | 644aa | ENSP00000359804 |
| FUBP1-214 | ENST00000703586 | 3330 | 666aa | ENSP00000515388 |
| FUBP1-204 | ENST00000421641 | 3426 | 687aa | ENSP00000402630 |
| FUBP1-201 | ENST00000294623 | 2201 | 653aa | ENSP00000294623 |
| FUBP1-215 | ENST00000703600 | 576 | 71aa | ENSP00000515389 |
| FUBP1-202 | ENST00000370767 | 2524 | 655aa | ENSP00000359803 |
| FUBP1-209 | ENST00000487684 | 535 | No protein | - |
| FUBP1-205 | ENST00000470287 | 726 | No protein | - |
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
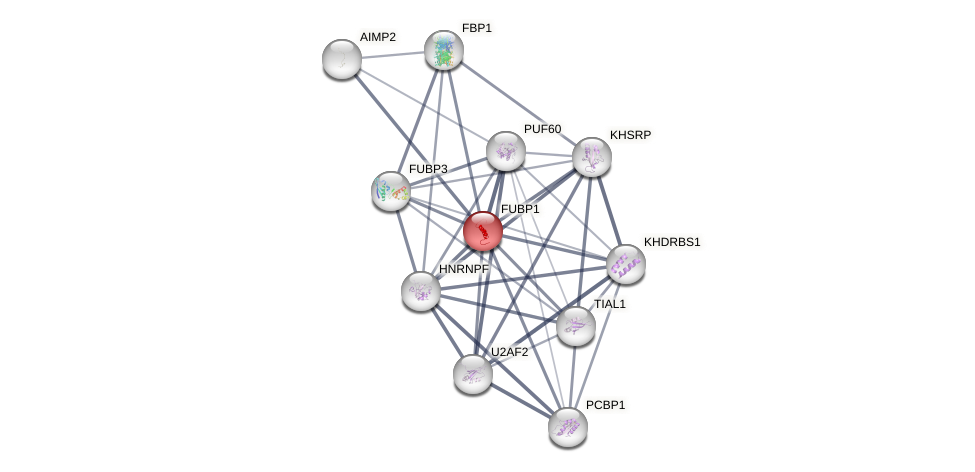
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000162613 | homo_sapiens | ENSP00000290341 | 33.2756 | 18.7175 | homo_sapiens | ENSP00000359804 | 29.8137 | 16.7702 |
| ENSG00000162613 | homo_sapiens | ENSP00000417196 | 39.206 | 23.3251 | homo_sapiens | ENSP00000359804 | 24.5342 | 14.5963 |
| ENSG00000162613 | homo_sapiens | ENSP00000263257 | 37.6016 | 22.9675 | homo_sapiens | ENSP00000359804 | 28.7267 | 17.5466 |
| ENSG00000162613 | homo_sapiens | ENSP00000505796 | 39.8922 | 25.3369 | homo_sapiens | ENSP00000359804 | 22.9814 | 14.5963 |
| ENSG00000162613 | homo_sapiens | ENSP00000365439 | 39.6552 | 23.7069 | homo_sapiens | ENSP00000359804 | 28.5714 | 17.0807 |
| ENSG00000162613 | homo_sapiens | ENSP00000448762 | 41.1602 | 25.1381 | homo_sapiens | ENSP00000359804 | 23.1366 | 14.1304 |
| ENSG00000162613 | homo_sapiens | ENSP00000371634 | 33.7229 | 18.5309 | homo_sapiens | ENSP00000359804 | 31.3665 | 17.236 |
| ENSG00000162613 | homo_sapiens | ENSP00000258729 | 33.1606 | 18.8256 | homo_sapiens | ENSP00000359804 | 29.8137 | 16.9255 |
| ENSG00000162613 | homo_sapiens | ENSP00000318177 | 70.8042 | 59.4406 | homo_sapiens | ENSP00000359804 | 62.8882 | 52.795 |
| ENSG00000162613 | homo_sapiens | ENSP00000305556 | 40.1685 | 25.2809 | homo_sapiens | ENSP00000359804 | 22.205 | 13.9752 |
| ENSG00000162613 | homo_sapiens | ENSP00000438875 | 38.6588 | 23.4714 | homo_sapiens | ENSP00000359804 | 30.4348 | 18.4783 |
| ENSG00000162613 | homo_sapiens | ENSP00000471146 | 64.7925 | 57.162 | homo_sapiens | ENSP00000359804 | 75.1553 | 66.3043 |
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 96.118 | 95.9627 | nomascus_leucogenys | ENSNLEP00000011932 | 94.0729 | 93.921 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 99.8447 | 99.8447 | pongo_abelii | ENSPPYP00000034882 | 96.8373 | 96.8373 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 67.0807 | 66.4596 | notamacropus_eugenii | ENSMEUP00000007246 | 70.2439 | 69.5935 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 99.2236 | 99.0683 | choloepus_hoffmanni | ENSCHOP00000007233 | 97.5573 | 97.4046 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 59.0062 | 52.9503 | dasypus_novemcinctus | ENSDNOP00000020240 | 74.0741 | 66.4717 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 85.7143 | 85.559 | erinaceus_europaeus | ENSEEUP00000001156 | 84.2748 | 84.1221 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 59.472 | 59.1615 | echinops_telfairi | ENSETEP00000000770 | 59.0139 | 58.7057 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 36.646 | 32.9193 | echinops_telfairi | ENSETEP00000013412 | 46.3654 | 41.6503 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 99.5342 | 99.3789 | callithrix_jacchus | ENSCJAP00000074153 | 96.2462 | 96.0961 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 99.8447 | 99.8447 | cercocebus_atys | ENSCATP00000038943 | 98.1679 | 98.1679 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 99.5342 | 99.5342 | macaca_fascicularis | ENSMFAP00000018131 | 96.102 | 96.102 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 99.8447 | 99.8447 | macaca_mulatta | ENSMMUP00000060500 | 89.9301 | 89.9301 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 99.6894 | 99.6894 | macaca_nemestrina | ENSMNEP00000045470 | 94.1349 | 94.1349 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 99.5342 | 99.5342 | papio_anubis | ENSPANP00000048843 | 96.102 | 96.102 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 99.6894 | 99.5342 | tursiops_truncatus | ENSTTRP00000000088 | 98.0153 | 97.8626 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 98.1366 | 97.9814 | mus_spretus | MGP_SPRETEiJ_P0063482 | 96.6361 | 96.4832 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 99.3789 | 99.2236 | loxodonta_africana | ENSLAFP00000002720 | 94.9555 | 94.8071 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 99.6894 | 99.3789 | bos_indicus_hybrid | ENSBIXP00005024266 | 95.1111 | 94.8148 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 99.8447 | 99.6894 | panthera_pardus | ENSPPRP00000004204 | 98.1679 | 98.0153 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 99.8447 | 99.6894 | marmota_marmota_marmota | ENSMMMP00000001878 | 97.7204 | 97.5684 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 99.6894 | 99.5342 | balaenoptera_musculus | ENSBMSP00010012527 | 96.6867 | 96.5361 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 99.2236 | 98.913 | cavia_porcellus | ENSCPOP00000002503 | 97.1125 | 96.8085 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 85.8696 | 84.0062 | monodelphis_domestica | ENSMODP00000045846 | 75.7534 | 74.1096 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 99.6894 | 99.5342 | bison_bison_bison | ENSBBBP00000003143 | 98.0153 | 97.8626 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 99.6894 | 99.5342 | sus_scrofa | ENSSSCP00000004075 | 96.6867 | 96.5361 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 99.3789 | 99.0683 | octodon_degus | ENSODEP00000003389 | 97.7099 | 97.4046 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 69.4099 | 66.3043 | octodon_degus | ENSODEP00000004817 | 85.4685 | 81.6444 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 99.2236 | 98.6025 | jaculus_jaculus | ENSJJAP00000019000 | 98.9164 | 98.2972 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 99.6894 | 99.5342 | ovis_aries_rambouillet | ENSOARP00020024491 | 96.6867 | 96.5361 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 82.4534 | 82.2981 | ochotona_princeps | ENSOPRP00000001100 | 81.5668 | 81.4132 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 99.5342 | 99.3789 | physeter_catodon | ENSPCTP00005022892 | 97.8626 | 97.7099 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 98.2919 | 98.1366 | mus_caroli | MGP_CAROLIEiJ_P0061039 | 96.789 | 96.6361 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 98.1366 | 97.8261 | mus_musculus | ENSMUSP00000143354 | 96.6361 | 96.3303 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 99.5342 | 99.2236 | dipodomys_ordii | ENSDORP00000026158 | 97.8626 | 97.5573 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 99.8447 | 99.6894 | saimiri_boliviensis_boliviensis | ENSSBOP00000022118 | 98.1679 | 98.0153 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 99.6894 | 99.5342 | vicugna_pacos | ENSVPAP00000004367 | 98.7692 | 98.6154 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 100 | 100 | chlorocebus_sabaeus | ENSCSAP00000016616 | 100 | 100 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 62.2671 | 62.1118 | tupaia_belangeri | ENSTBEP00000015082 | 65.2033 | 65.0406 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 99.3789 | 99.0683 | mesocricetus_auratus | ENSMAUP00000017726 | 97.2644 | 96.9605 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 99.8447 | 99.6894 | phocoena_sinus | ENSPSNP00000003290 | 96.6917 | 96.5414 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 92.0807 | 91.3043 | carlito_syrichta | ENSTSYP00000027361 | 90.8116 | 90.0459 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 99.8447 | 99.6894 | panthera_tigris_altaica | ENSPTIP00000022774 | 98.1679 | 98.0153 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 98.4472 | 98.2919 | rattus_norvegicus | ENSRNOP00000077531 | 93.9259 | 93.7778 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 99.6894 | 99.5342 | prolemur_simus | ENSPSMP00000021502 | 98.0153 | 97.8626 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 99.0683 | 98.913 | microtus_ochrogaster | ENSMOCP00000015612 | 97.5535 | 97.4006 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 83.5404 | 83.3851 | sorex_araneus | ENSSARP00000010616 | 82.1374 | 81.9847 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 98.4472 | 98.1366 | peromyscus_maniculatus_bairdii | ENSPEMP00000025341 | 97.0904 | 96.7841 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 97.9814 | 97.8261 | mus_spicilegus | ENSMSIP00000028928 | 94.7447 | 94.5946 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 98.4472 | 98.1366 | cricetulus_griseus_chok1gshd | ENSCGRP00001005473 | 95.4819 | 95.1807 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 95.3416 | 95.1863 | pteropus_vampyrus | ENSPVAP00000013801 | 93.8838 | 93.7309 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 99.2236 | 99.0683 | myotis_lucifugus | ENSMLUP00000013476 | 95.5157 | 95.3662 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 98.1366 | 97.205 | vombatus_ursinus | ENSVURP00010014425 | 92.1283 | 91.2536 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 99.8447 | 99.5342 | rhinolophus_ferrumequinum | ENSRFEP00010009737 | 96.6917 | 96.391 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 83.6957 | 80.1242 | heterocephalus_glaber_female | ENSHGLP00000003814 | 84.4828 | 80.8777 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 99.5342 | 99.2236 | heterocephalus_glaber_female | ENSHGLP00000011740 | 95.9581 | 95.6587 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 98.2919 | 98.2919 | rhinopithecus_bieti | ENSRBIP00000039069 | 95.045 | 95.045 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 99.8447 | 99.6894 | sciurus_vulgaris | ENSSVLP00005027631 | 98.1679 | 98.0153 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 99.2236 | 98.913 | procavia_capensis | ENSPCAP00000011822 | 97.7064 | 97.4006 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 95.1863 | 95.0311 | nannospalax_galili | ENSNGAP00000012459 | 97.6115 | 97.4522 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 99.5342 | 99.2236 | chinchilla_lanigera | ENSCLAP00000008019 | 97.8626 | 97.5573 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 99.3789 | 99.2236 | cervus_hanglu_yarkandensis | ENSCHYP00000039126 | 95.952 | 95.8021 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 99.3789 | 99.2236 | catagonus_wagneri | ENSCWAP00000006129 | 97.7099 | 97.5573 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 99.8447 | 99.6894 | ictidomys_tridecemlineatus | ENSSTOP00000029681 | 97.7204 | 97.5684 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 100 | 99.8447 | otolemur_garnettii | ENSOGAP00000012762 | 95.9762 | 95.8271 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 81.8323 | 81.8323 | cebus_imitator | ENSCCAP00000037356 | 97.7737 | 97.7737 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 96.118 | 95.9627 | urocitellus_parryii | ENSUPAP00010019690 | 97.7883 | 97.6303 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 98.2919 | 98.2919 | rhinopithecus_roxellana | ENSRROP00000040842 | 95.045 | 95.045 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 98.7578 | 98.6025 | mus_pahari | MGP_PahariEiJ_P0071971 | 97.0992 | 96.9466 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 99.8447 | 99.6894 | propithecus_coquereli | ENSPCOP00000025555 | 98.1679 | 98.0153 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 17.7019 | 11.1801 | caenorhabditis_elegans | Y119D3B.17.2 | 26.3889 | 16.6667 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 20.4969 | 12.1118 | caenorhabditis_elegans | Y59A8B.10d.1 | 31.6547 | 18.705 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 41.4596 | 26.8634 | caenorhabditis_elegans | C36E6.1b.1 | 41.6537 | 26.9891 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 40.6832 | 24.8447 | caenorhabditis_elegans | M01A10.1.1 | 44.7099 | 27.3038 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 23.6025 | 13.6646 | caenorhabditis_elegans | M88.5c.1 | 17.7986 | 10.3045 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 25.1553 | 16.1491 | drosophila_melanogaster | FBpp0293521 | 28.2723 | 18.1501 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 25.6211 | 15.2174 | drosophila_melanogaster | FBpp0306419 | 21.1538 | 12.5641 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 23.2919 | 14.2857 | drosophila_melanogaster | FBpp0293680 | 23.511 | 14.4201 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 18.0124 | 11.4907 | ciona_intestinalis | ENSCINP00000001984 | 32.493 | 20.7283 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 15.528 | 9.16149 | ciona_intestinalis | ENSCINP00000025950 | 35.2113 | 20.7746 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 27.4845 | 14.1304 | ciona_intestinalis | ENSCINP00000008726 | 29.9492 | 15.3976 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 37.2671 | 31.2112 | petromyzon_marinus | ENSPMAP00000009129 | 74.7664 | 62.6168 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 27.9503 | 22.9814 | petromyzon_marinus | ENSPMAP00000001804 | 72.8745 | 59.919 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 59.3168 | 49.2236 | petromyzon_marinus | ENSPMAP00000007628 | 63.1405 | 52.3967 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 57.4534 | 46.118 | eptatretus_burgeri | ENSEBUP00000026225 | 59.4855 | 47.7492 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 80.7453 | 75.7764 | callorhinchus_milii | ENSCMIP00000020501 | 77.6119 | 72.8358 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 83.8509 | 77.1739 | lepisosteus_oculatus | ENSLOCP00000004486 | 78.4884 | 72.2384 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 82.764 | 77.3292 | clupea_harengus | ENSCHAP00000021630 | 76.912 | 71.8615 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 82.2981 | 75.6211 | danio_rerio | ENSDARP00000075883 | 82.5545 | 75.8567 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 76.2422 | 69.4099 | carassius_auratus | ENSCARP00000113105 | 75.0765 | 68.3486 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 81.2112 | 74.0683 | carassius_auratus | ENSCARP00000070134 | 71.7421 | 65.4321 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 70.6522 | 65.8385 | astyanax_mexicanus | ENSAMXP00000005663 | 85.6874 | 79.8493 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 82.2981 | 76.087 | ictalurus_punctatus | ENSIPUP00000014563 | 81.1639 | 75.0383 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 82.6087 | 75.4658 | esox_lucius | ENSELUP00000007201 | 80.3625 | 73.4139 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 81.8323 | 74.2236 | oncorhynchus_kisutch | ENSOKIP00005094288 | 79.9697 | 72.5341 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 81.9876 | 74.5342 | oncorhynchus_kisutch | ENSOKIP00005063771 | 78.9238 | 71.7489 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 81.9876 | 74.5342 | oncorhynchus_mykiss | ENSOMYP00000116650 | 76.7442 | 69.7674 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 48.2919 | 39.5963 | oncorhynchus_mykiss | ENSOMYP00000141605 | 29.3673 | 24.0793 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 81.677 | 74.2236 | salmo_salar | ENSSSAP00000036874 | 75.5747 | 68.6782 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 81.8323 | 74.8447 | salmo_salar | ENSSSAP00000123379 | 78.8922 | 72.1557 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 81.677 | 74.5342 | salmo_trutta | ENSSTUP00000054565 | 78.7425 | 71.8563 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 79.0373 | 71.7391 | salmo_trutta | ENSSTUP00000086669 | 76.6566 | 69.5783 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 78.5714 | 70.3416 | gadus_morhua | ENSGMOP00000045224 | 78.0864 | 69.9074 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 82.6087 | 76.7081 | fundulus_heteroclitus | ENSFHEP00000004232 | 81.3456 | 75.5352 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 82.6087 | 76.5528 | poecilia_reticulata | ENSPREP00000026516 | 81.0976 | 75.1524 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 77.6398 | 71.5839 | xiphophorus_maculatus | ENSXMAP00000007278 | 83.3333 | 76.8333 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 79.5031 | 73.2919 | oryzias_latipes | ENSORLP00000004413 | 83.7971 | 77.2504 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 83.0745 | 76.2422 | cyclopterus_lumpus | ENSCLMP00005050896 | 76.3195 | 70.0428 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 82.764 | 76.7081 | oreochromis_niloticus | ENSONIP00000023141 | 79.4337 | 73.6215 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 82.764 | 76.7081 | haplochromis_burtoni | ENSHBUP00000008348 | 79.4337 | 73.6215 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 82.9193 | 76.7081 | astatotilapia_calliptera | ENSACLP00000009848 | 79.5827 | 73.6215 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 83.5404 | 77.1739 | sparus_aurata | ENSSAUP00010022111 | 77.2989 | 71.408 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 83.5404 | 77.4845 | lates_calcarifer | ENSLCAP00010054609 | 79.2342 | 73.4904 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 93.7888 | 93.0124 | sphenodon_punctatus | ENSSPUP00000010900 | 87.4096 | 86.686 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 60.0932 | 58.6957 | laticauda_laticaudata | ENSLLTP00000003806 | 87.9545 | 85.9091 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 93.7888 | 92.3913 | notechis_scutatus | ENSNSUP00000008940 | 90.8271 | 89.4737 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 95.4969 | 94.0994 | pseudonaja_textilis | ENSPTXP00000008783 | 90.176 | 88.8563 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 91.3043 | 89.9068 | anas_platyrhynchos_platyrhynchos | ENSAPLP00000005641 | 95.7655 | 94.2997 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 97.3602 | 96.5839 | gallus_gallus | ENSGALP00010032622 | 91.5328 | 90.8029 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 85.7143 | 82.9193 | meleagris_gallopavo | ENSMGAP00000026692 | 64.5614 | 62.4561 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 96.8944 | 95.8075 | serinus_canaria | ENSSCAP00000019921 | 96.7442 | 95.6589 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 97.0497 | 95.9627 | parus_major | ENSPMJP00000026673 | 94.5537 | 93.4947 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 88.5093 | 86.8012 | anolis_carolinensis | ENSACAP00000033463 | 83.8235 | 82.2059 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 79.8137 | 73.7578 | neolamprologus_brichardi | ENSNBRP00000019630 | 86.0972 | 79.5645 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 95.4969 | 94.0994 | naja_naja | ENSNNAP00000002231 | 86.9873 | 85.7143 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 95.4969 | 93.9441 | podarcis_muralis | ENSPMRP00000015041 | 86.9873 | 85.5728 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 89.441 | 88.9752 | geospiza_fortis | ENSGFOP00000021056 | 94.2717 | 93.7807 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 96.8944 | 95.8075 | taeniopygia_guttata | ENSTGUP00000028480 | 96.7442 | 95.6589 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 86.1801 | 80.7453 | erpetoichthys_calabaricus | ENSECRP00000010190 | 83.2084 | 77.961 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 82.9193 | 76.8634 | kryptolebias_marmoratus | ENSKMAP00000025624 | 81.155 | 75.228 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 95.6522 | 94.0994 | salvator_merianae | ENSSMRP00000026146 | 89.0173 | 87.5723 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 82.764 | 76.5528 | oryzias_javanicus | ENSOJAP00000029425 | 77.8102 | 71.9708 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 79.1925 | 73.7578 | amphilophus_citrinellus | ENSACIP00000025872 | 82.1256 | 76.4895 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 82.9193 | 76.3975 | dicentrarchus_labrax | ENSDLAP00005062682 | 82.5348 | 76.0433 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 80.1242 | 74.0683 | amphiprion_percula | ENSAPEP00000017167 | 84.5902 | 78.1967 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 82.1429 | 76.087 | oryzias_sinensis | ENSOSIP00000043033 | 75.4636 | 69.9001 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 91.6149 | 90.9938 | pelodiscus_sinensis | ENSPSIP00000014601 | 96.7213 | 96.0656 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 83.2298 | 77.0186 | seriola_dumerili | ENSSDUP00000010172 | 76.5714 | 70.8571 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 80.9006 | 74.8447 | cottoperca_gobio | ENSCGOP00000006768 | 82.0472 | 75.9055 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 92.3913 | 91.1491 | struthio_camelus_australis | ENSSCUP00000011494 | 91.3979 | 90.169 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 97.205 | 96.2733 | anser_brachyrhynchus | ENSABRP00000021452 | 94.705 | 93.7973 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 81.9876 | 75.7764 | gasterosteus_aculeatus | ENSGACP00000021873 | 82.6291 | 76.3693 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 82.2981 | 75.9317 | cynoglossus_semilaevis | ENSCSEP00000033371 | 82.8125 | 76.4062 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 82.9193 | 77.0186 | poecilia_formosa | ENSPFOP00000000319 | 81.6514 | 75.841 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 82.764 | 76.5528 | myripristis_murdjan | ENSMMDP00005047186 | 77.5837 | 71.7613 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 83.5404 | 76.8634 | sander_lucioperca | ENSSLUP00000001639 | 80.2985 | 73.8806 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 97.0497 | 96.2733 | strigops_habroptila | ENSSHBP00005016263 | 92.1829 | 91.4454 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 96.8944 | 95.9627 | terrapene_carolina_triunguis | ENSTMTP00000011103 | 94.2598 | 93.3535 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 91.4596 | 90.3727 | terrapene_carolina_triunguis | ENSTMTP00000022543 | 90.6154 | 89.5385 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 8.85093 | 7.76398 | hucho_hucho | ENSHHUP00000018041 | 64.7727 | 56.8182 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 29.3478 | 24.8447 | hucho_hucho | ENSHHUP00000018048 | 83.6283 | 70.7965 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 11.1801 | 9.78261 | hucho_hucho | ENSHHUP00000005147 | 81.8182 | 71.5909 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 12.1118 | 11.0248 | hucho_hucho | ENSHHUP00000005156 | 73.5849 | 66.9811 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 83.0745 | 76.3975 | betta_splendens | ENSBSLP00000051099 | 80.2099 | 73.7631 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 83.3851 | 77.3292 | pygocentrus_nattereri | ENSPNAP00000010608 | 81.6109 | 75.6839 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 83.2298 | 76.8634 | anabas_testudineus | ENSATEP00000026957 | 80.6015 | 74.4361 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 13.8199 | 8.54037 | ciona_savignyi | ENSCSAVP00000004755 | 42.381 | 26.1905 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 9.31677 | 5.59006 | ciona_savignyi | ENSCSAVP00000007214 | 49.1803 | 29.5082 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 27.6398 | 14.1304 | ciona_savignyi | ENSCSAVP00000018813 | 30.6897 | 15.6897 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 83.5404 | 77.3292 | seriola_lalandi_dorsalis | ENSSLDP00000029697 | 79.1176 | 73.2353 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 83.3851 | 76.5528 | labrus_bergylta | ENSLBEP00000016520 | 77.8261 | 71.4493 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 83.2298 | 77.0186 | pundamilia_nyererei | ENSPNYP00000005930 | 82.3349 | 76.1905 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 83.5404 | 77.6398 | mastacembelus_armatus | ENSMAMP00000011042 | 81.1463 | 75.4148 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 83.0745 | 76.8634 | stegastes_partitus | ENSSPAP00000013419 | 81.5549 | 75.4573 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 58.2298 | 47.205 | oncorhynchus_tshawytscha | ENSOTSP00005079837 | 57.1646 | 46.3415 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 77.4845 | 69.7205 | oncorhynchus_tshawytscha | ENSOTSP00005052380 | 75.6061 | 68.0303 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 79.5031 | 72.5155 | acanthochromis_polyacanthus | ENSAPOP00000029133 | 75.405 | 68.7776 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 97.3602 | 96.5839 | aquila_chrysaetos_chrysaetos | ENSACCP00020005947 | 97.6636 | 96.8847 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 82.4534 | 76.087 | nothobranchius_furzeri | ENSNFUP00015022408 | 77.6316 | 71.6374 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 82.4534 | 77.0186 | cyprinodon_variegatus | ENSCVAP00000011980 | 80.9451 | 75.6098 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 83.3851 | 76.7081 | larimichthys_crocea | ENSLCRP00005060617 | 73.461 | 67.5787 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 82.9193 | 75.4658 | takifugu_rubripes | ENSTRUP00000010975 | 79.4643 | 72.3214 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 97.0497 | 95.9627 | ficedula_albicollis | ENSFALP00000030383 | 94.697 | 93.6364 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 83.0745 | 75.9317 | hippocampus_comes | ENSHCOP00000000166 | 80.0898 | 73.2036 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 97.0497 | 96.2733 | coturnix_japonica | ENSCJPP00005009616 | 96.7492 | 95.9752 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 81.2112 | 75.4658 | poecilia_latipinna | ENSPLAP00000030204 | 81.5913 | 75.819 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 57.4534 | 50 | sinocyclocheilus_grahami | ENSSGRP00000020813 | 73.7052 | 64.1434 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 82.764 | 74.5342 | sinocyclocheilus_grahami | ENSSGRP00000016742 | 80.7576 | 72.7273 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 83.2298 | 77.0186 | maylandia_zebra | ENSMZEP00005032611 | 82.3349 | 76.1905 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 22.6708 | 20.0311 | tetraodon_nigroviridis | ENSTNIP00000023082 | 86.3905 | 76.3314 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 24.8447 | 22.205 | tetraodon_nigroviridis | ENSTNIP00000006595 | 59.9251 | 53.5581 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 81.9876 | 75.9317 | scleropages_formosus | ENSSFOP00015041261 | 79.2793 | 73.4234 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 83.3851 | 77.0186 | scophthalmus_maximus | ENSSMAP00000019597 | 75.8475 | 70.0565 |
| ENSG00000162613 | homo_sapiens | ENSP00000359804 | 82.2981 | 76.087 | oryzias_melastigma | ENSOMEP00000020091 | 78.7519 | 72.8083 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0003697 | enables single-stranded DNA binding | 1 | TAS | Homo_sapiens(9606) | Function |
| GO:0003723 | enables RNA binding | 1 | HDA | Homo_sapiens(9606) | Function |
| GO:0003729 | enables mRNA binding | 1 | IBA | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005634 | is_active_in nucleus | 2 | IBA,IDA | Homo_sapiens(9606) | Component |
| GO:0005654 | located_in nucleoplasm | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005737 | is_active_in cytoplasm | 1 | IBA | Homo_sapiens(9606) | Component |
| GO:0010468 | involved_in regulation of gene expression | 2 | IBA,TAS | Homo_sapiens(9606) | Process |
| GO:0010628 | involved_in positive regulation of gene expression | 1 | IDA | Homo_sapiens(9606) | Process |