Welcome to
RBP World!
Description
| Ensembl ID | ENSG00000141510 | Gene ID | 7157 | Accession | 11998 |
| Symbol | TP53 | Alias | P53;BCC7;LFS1;BMFS5;TRP53 | Full Name | tumor protein p53 |
| Status | Confidence | Length | 25760 bases | Strand | Minus strand |
| Position | 17 : 7661779 - 7687538 | RNA binding domain | N.A. | ||
| Summary | This gene encodes a tumor suppressor protein containing transcriptional activation, DNA binding, and oligomerization domains. The encoded protein responds to diverse cellular stresses to regulate expression of target genes, thereby inducing cell cycle arrest, apoptosis, senescence, DNA repair, or changes in metabolism. Mutations in this gene are associated with a variety of human cancers, including hereditary cancers such as Li-Fraumeni syndrome. Alternative splicing of this gene and the use of alternate promoters result in multiple transcript variants and isoforms. Additional isoforms have also been shown to result from the use of alternate translation initiation codons from identical transcript variants (PMIDs: 12032546, 20937277). [provided by RefSeq, Dec 2016] | ||||
RNA binding domains (RBDs)
| Protein | Domain | Pfam ID | E-value | Domain number |
|---|
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| TP53-203 | ENST00000413465 | 1018 | 285aa | ENSP00000410739 |
| TP53-227 | ENST00000635293 | 1883 | 410aa | ENSP00000488924 |
| TP53-202 | ENST00000359597 | 1152 | 343aa | ENSP00000352610 |
| TP53-208 | ENST00000504290 | 2331 | 214aa | ENSP00000484409 |
| TP53-209 | ENST00000504937 | 2271 | 261aa | ENSP00000481179 |
| TP53-213 | ENST00000510385 | 2404 | 209aa | ENSP00000478499 |
| TP53-221 | ENST00000610623 | 2331 | 187aa | ENSP00000477531 |
| TP53-222 | ENST00000618944 | 2404 | 182aa | ENSP00000481401 |
| TP53-223 | ENST00000619186 | 2271 | 234aa | ENSP00000484375 |
| TP53-219 | ENST00000610292 | 2639 | 354aa | ENSP00000478219 |
| TP53-225 | ENST00000620739 | 2579 | 354aa | ENSP00000481638 |
| TP53-204 | ENST00000420246 | 2653 | 341aa | ENSP00000391127 |
| TP53-206 | ENST00000455263 | 2580 | 346aa | ENSP00000398846 |
| TP53-220 | ENST00000610538 | 2580 | 307aa | ENSP00000480868 |
| TP53-226 | ENST00000622645 | 2653 | 302aa | ENSP00000482222 |
| TP53-211 | ENST00000508793 | 2660 | 393aa | ENSP00000424104 |
| TP53-207 | ENST00000503591 | 2552 | 393aa | ENSP00000426252 |
| TP53-214 | ENST00000514944 | 2170 | 300aa | ENSP00000423862 |
| TP53-205 | ENST00000445888 | 2506 | 393aa | ENSP00000391478 |
| TP53-212 | ENST00000509690 | 2106 | 261aa | ENSP00000425104 |
| TP53-218 | ENST00000604348 | 2488 | 386aa | ENSP00000473895 |
| TP53-224 | ENST00000619485 | 2506 | 354aa | ENSP00000482537 |
| TP53-201 | ENST00000269305 | 2512 | 393aa | ENSP00000269305 |
| TP53-217 | ENST00000576024 | 175 | 31aa | ENSP00000458393 |
| TP53-216 | ENST00000574684 | 104 | No protein | - |
| TP53-210 | ENST00000505014 | 1261 | No protein | - |
| TP53-215 | ENST00000571370 | 2275 | No protein | - |
Phenotypes
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| 64622 | Glioblastoma | 4E-12 | 26424050 |
| 66549 | Carcinoma, Basal Cell | 2E-20 | 21946351 |
| 70786 | Carcinoma, Basal Cell | 2E-10 | 27539887 |
| 79315 | Carcinoma, Basal Cell | 1E-20 | 25855136 |
| 88646 | Hormones | 1E-15 | 26014426 |
| 88647 | Sex Hormone-Binding Globulin | 1E-15 | 26014426 |
| 95054 | Carcinoma, Basal Cell | 4E-22 | 24403052 |
| 100295 | Sex Hormone-Binding Globulin | 2E-21 | 22829776 |
| 104807 | Glioma | 9E-20 | 26424050 |
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
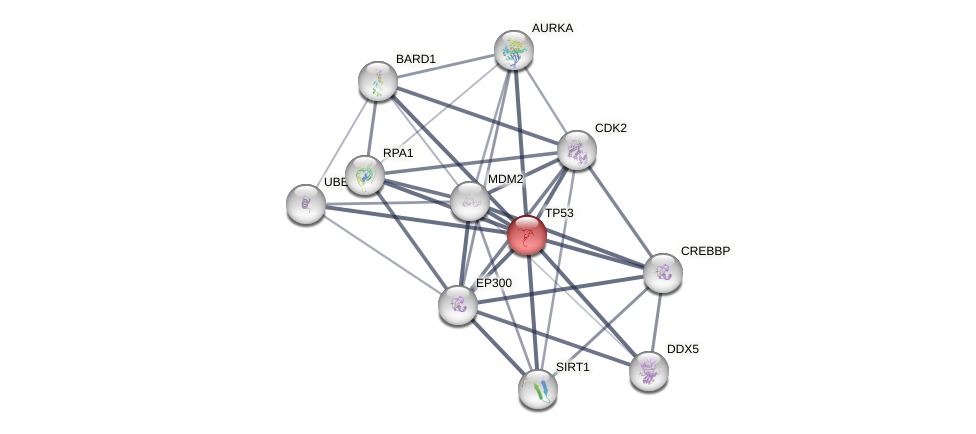
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000141510 | homo_sapiens | ENSP00000367545 | 36.6352 | 26.4151 | homo_sapiens | ENSP00000269305 | 59.2875 | 42.7481 |
| ENSG00000141510 | homo_sapiens | ENSP00000264731 | 33.5294 | 22.6471 | homo_sapiens | ENSP00000269305 | 58.0153 | 39.1857 |
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 89.8219 | 88.8041 | nomascus_leucogenys | ENSNLEP00000011861 | 97.7839 | 96.6759 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 100 | 100 | pan_paniscus | ENSPPAP00000011040 | 100 | 100 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 98.2188 | 97.7099 | pongo_abelii | ENSPPYP00000045316 | 92.3445 | 91.866 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 100 | 100 | pan_troglodytes | ENSPTRP00000014836 | 100 | 100 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 83.9695 | 83.715 | gorilla_gorilla | ENSGGOP00000030954 | 81.8859 | 81.6377 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 70.9924 | 59.0331 | ornithorhynchus_anatinus | ENSOANP00000037616 | 59.6154 | 49.5727 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 72.7735 | 65.1399 | sarcophilus_harrisii | ENSSHAP00000033581 | 68.4211 | 61.244 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 49.1094 | 45.5471 | notamacropus_eugenii | ENSMEUP00000011978 | 85.022 | 78.8546 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 84.9873 | 80.4071 | dasypus_novemcinctus | ENSDNOP00000004075 | 86.5285 | 81.8653 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 72.0102 | 63.3588 | erinaceus_europaeus | ENSEEUP00000013295 | 75.4667 | 66.4 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 46.3104 | 41.2214 | echinops_telfairi | ENSETEP00000008111 | 55.3191 | 49.2401 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 62.341 | 57.7608 | echinops_telfairi | ENSETEP00000007296 | 73.1343 | 67.7612 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 90.3308 | 86.514 | callithrix_jacchus | ENSCJAP00000066800 | 89.6465 | 85.8586 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 93.8931 | 92.112 | cercocebus_atys | ENSCATP00000032045 | 91.791 | 90.0498 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 97.201 | 95.6743 | macaca_fascicularis | ENSMFAP00000038526 | 97.201 | 95.6743 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 97.201 | 95.6743 | macaca_mulatta | ENSMMUP00000011334 | 88.2217 | 86.836 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 56.4885 | 52.6718 | macaca_nemestrina | ENSMNEP00000030865 | 82.2222 | 76.6667 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 96.9466 | 95.6743 | papio_anubis | ENSPANP00000019643 | 96.9466 | 95.6743 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 93.8931 | 92.112 | mandrillus_leucophaeus | ENSMLEP00000037099 | 91.791 | 90.0498 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 89.5674 | 85.2417 | tursiops_truncatus | ENSTTRP00000009721 | 90.9561 | 86.5633 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 86.2595 | 80.4071 | vulpes_vulpes | ENSVVUP00000039187 | 88.9764 | 82.9396 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 83.2061 | 77.3537 | mus_spretus | MGP_SPRETEiJ_P0028933 | 83.8462 | 77.9487 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 87.5318 | 83.4606 | equus_asinus | ENSEASP00005028399 | 90.2887 | 86.0892 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 85.4962 | 79.8982 | capra_hircus | ENSCHIP00000030141 | 87.9581 | 82.199 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 65.6489 | 58.2697 | loxodonta_africana | ENSLAFP00000028793 | 70.2997 | 62.3978 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 68.7023 | 62.341 | loxodonta_africana | ENSLAFP00000023389 | 74.1758 | 67.3077 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 68.4478 | 63.3588 | loxodonta_africana | ENSLAFP00000023554 | 73.297 | 67.8474 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 65.1399 | 59.0331 | loxodonta_africana | ENSLAFP00000027515 | 71.1111 | 64.4444 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 65.9033 | 60.0509 | loxodonta_africana | ENSLAFP00000027808 | 71.9444 | 65.5556 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 64.1221 | 56.9975 | loxodonta_africana | ENSLAFP00000028173 | 69.0411 | 61.3699 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 82.1883 | 78.626 | loxodonta_africana | ENSLAFP00000006292 | 82.398 | 78.8265 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 67.1756 | 61.3232 | loxodonta_africana | ENSLAFP00000025852 | 72.9282 | 66.5746 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 52.6718 | 48.0916 | loxodonta_africana | ENSLAFP00000027441 | 71.875 | 65.625 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 64.631 | 57.7608 | loxodonta_africana | ENSLAFP00000018379 | 69.9725 | 62.5344 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 65.6489 | 59.2875 | loxodonta_africana | ENSLAFP00000027253 | 70.2997 | 63.4877 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 68.1934 | 62.341 | loxodonta_africana | ENSLAFP00000024998 | 73.6264 | 67.3077 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 86.2595 | 80.6616 | panthera_leo | ENSPLOP00000027743 | 87.8238 | 82.1244 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 87.5318 | 80.6616 | ailuropoda_melanoleuca | ENSAMEP00000017825 | 72.5738 | 66.8776 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 85.7506 | 80.1527 | bos_indicus_hybrid | ENSBIXP00005039643 | 67.5351 | 63.1263 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 90.3308 | 85.7506 | oryctolagus_cuniculus | ENSOCUP00000000989 | 90.7928 | 86.1893 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 87.7863 | 83.715 | equus_caballus | ENSECAP00000074678 | 80.9859 | 77.23 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 86.514 | 80.6616 | canis_lupus_familiaris | ENSCAFP00845034138 | 89.2388 | 83.2021 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 86.2595 | 80.6616 | panthera_pardus | ENSPPRP00000022844 | 87.8238 | 82.1244 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 91.0941 | 86.514 | marmota_marmota_marmota | ENSMMMP00000022491 | 91.3265 | 86.7347 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 89.313 | 85.4962 | balaenoptera_musculus | ENSBMSP00010020707 | 90.6977 | 86.8217 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 83.715 | 78.1171 | cavia_porcellus | ENSCPOP00000029751 | 77.7778 | 72.5768 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 86.0051 | 80.4071 | felis_catus | ENSFCAP00000045749 | 65.251 | 61.0039 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 87.0229 | 80.4071 | ursus_americanus | ENSUAMP00000000913 | 89.7638 | 82.9396 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 86.0051 | 80.4071 | bison_bison_bison | ENSBBBP00000018891 | 87.5648 | 81.8653 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 89.5674 | 85.7506 | monodon_monoceros | ENSMMNP00015021985 | 90.9561 | 87.0801 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 85.4962 | 80.4071 | sus_scrofa | ENSSSCP00000050664 | 85.0633 | 80 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 88.2952 | 86.0051 | microcebus_murinus | ENSMICP00000039812 | 90.1299 | 87.7922 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 83.715 | 79.8982 | octodon_degus | ENSODEP00000024162 | 84.7938 | 80.9278 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 84.2239 | 78.8804 | jaculus_jaculus | ENSJJAP00000008090 | 88.9785 | 83.3333 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 85.4962 | 79.6438 | ovis_aries_rambouillet | ENSOARP00020014679 | 87.9581 | 81.9372 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 87.0229 | 80.1527 | ursus_maritimus | ENSUMAP00000023207 | 89.7638 | 82.6772 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 82.9517 | 75.827 | ochotona_princeps | ENSOPRP00000005906 | 87.1658 | 79.6791 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 88.8041 | 84.7328 | delphinapterus_leucas | ENSDLEP00000029611 | 88.1313 | 84.0909 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 90.0763 | 85.7506 | physeter_catodon | ENSPCTP00005020186 | 91.4729 | 87.0801 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 83.715 | 77.6081 | mus_caroli | MGP_CAROLIEiJ_P0027554 | 84.359 | 78.2051 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 82.9517 | 77.3537 | mus_musculus | ENSMUSP00000104298 | 83.5897 | 77.9487 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 77.0992 | 72.0102 | dipodomys_ordii | ENSDORP00000023664 | 85.1124 | 79.4944 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 84.9873 | 78.626 | mustela_putorius_furo | ENSMPUP00000008945 | 89.0667 | 82.4 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 72.7735 | 69.4657 | aotus_nancymaae | ENSANAP00000020165 | 77.9292 | 74.3869 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 77.6081 | 72.2646 | aotus_nancymaae | ENSANAP00000025592 | 90.5044 | 84.273 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 69.9746 | 66.9211 | aotus_nancymaae | ENSANAP00000040175 | 83.8415 | 80.1829 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 85.7506 | 83.715 | saimiri_boliviensis_boliviensis | ENSSBOP00000008181 | 94.6629 | 92.4157 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 97.201 | 95.9288 | chlorocebus_sabaeus | ENSCSAP00000007551 | 97.201 | 95.9288 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 55.2163 | 51.145 | tupaia_belangeri | ENSTBEP00000010189 | 57.4074 | 53.1746 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 83.2061 | 76.0814 | mesocricetus_auratus | ENSMAUP00000025852 | 82.7848 | 75.6962 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 89.313 | 85.4962 | phocoena_sinus | ENSPSNP00000013453 | 90.6977 | 86.8217 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 88.5496 | 83.715 | camelus_dromedarius | ENSCDRP00005013165 | 91.3386 | 86.3517 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 91.8575 | 89.313 | carlito_syrichta | ENSTSYP00000023752 | 93.0412 | 90.4639 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 57.5064 | 52.4173 | panthera_tigris_altaica | ENSPTIP00000024834 | 75.5853 | 68.8963 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 83.9695 | 77.8626 | rattus_norvegicus | ENSRNOP00000092831 | 81.2808 | 75.3695 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 78.1171 | 74.8092 | prolemur_simus | ENSPSMP00000026875 | 76.9424 | 73.6842 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 83.2061 | 79.8982 | microtus_ochrogaster | ENSMOCP00000023219 | 86.0526 | 82.6316 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 84.2239 | 78.8804 | sorex_araneus | ENSSARP00000005616 | 85.7513 | 80.3109 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 85.7506 | 82.4427 | peromyscus_maniculatus_bairdii | ENSPEMP00000010300 | 86.8557 | 83.5052 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 86.0051 | 80.4071 | bos_mutus | ENSBMUP00000028863 | 87.5648 | 81.8653 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 52.1628 | 47.0738 | mus_spicilegus | ENSMSIP00000005017 | 78.8462 | 71.1538 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 82.9517 | 77.3537 | mus_spicilegus | ENSMSIP00000031034 | 84.0206 | 78.3505 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 83.4606 | 76.8448 | cricetulus_griseus_chok1gshd | ENSCGRP00001011268 | 83.4606 | 76.8448 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 63.1043 | 58.2697 | pteropus_vampyrus | ENSPVAP00000015400 | 63.4271 | 58.5678 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 80.4071 | 73.7914 | myotis_lucifugus | ENSMLUP00000005861 | 80.6122 | 73.9796 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 68.4478 | 62.8499 | myotis_lucifugus | ENSMLUP00000016733 | 69.3299 | 63.6598 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 74.3003 | 66.1578 | vombatus_ursinus | ENSVURP00010004263 | 78.9189 | 70.2703 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 86.2595 | 80.916 | rhinolophus_ferrumequinum | ENSRFEP00010016808 | 89.2105 | 83.6842 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 85.4962 | 79.1349 | neovison_vison | ENSNVIP00000005141 | 88.6544 | 82.058 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 85.7506 | 80.1527 | bos_taurus | ENSBTAP00000001420 | 87.3057 | 81.6062 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 86.2595 | 82.4427 | heterocephalus_glaber_female | ENSHGLP00000054376 | 86.9231 | 83.0769 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 97.4555 | 96.1832 | rhinopithecus_bieti | ENSRBIP00000028632 | 97.4555 | 96.1832 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 86.514 | 80.6616 | canis_lupus_dingo | ENSCAFP00020009510 | 89.2388 | 83.2021 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 89.313 | 86.2595 | sciurus_vulgaris | ENSSVLP00005019717 | 90 | 86.9231 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 74.3003 | 66.6667 | phascolarctos_cinereus | ENSPCIP00000039513 | 58.0517 | 52.0875 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 79.1349 | 74.0458 | procavia_capensis | ENSPCAP00000011892 | 80.7792 | 75.5844 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 89.0585 | 84.4784 | nannospalax_galili | ENSNGAP00000007306 | 89.5141 | 84.9105 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 86.0051 | 80.4071 | bos_grunniens | ENSBGRP00000021669 | 87.5648 | 81.8653 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 86.7684 | 82.6972 | chinchilla_lanigera | ENSCLAP00000010404 | 87.2123 | 83.1202 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 86.0051 | 79.6438 | cervus_hanglu_yarkandensis | ENSCHYP00000017281 | 86.8895 | 80.4627 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 87.7863 | 83.715 | catagonus_wagneri | ENSCWAP00000005800 | 89.3782 | 85.2332 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 90.8397 | 86.2595 | ictidomys_tridecemlineatus | ENSSTOP00000019678 | 91.3043 | 86.7008 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 87.7863 | 83.9695 | otolemur_garnettii | ENSOGAP00000015106 | 87.7863 | 83.9695 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 93.3842 | 90.5852 | cebus_imitator | ENSCCAP00000029525 | 92.9114 | 90.1266 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 90.8397 | 86.0051 | urocitellus_parryii | ENSUPAP00010019485 | 90.6091 | 85.7868 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 86.7684 | 81.4249 | moschus_moschiferus | ENSMMSP00000026578 | 88.1137 | 82.6873 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 97.4555 | 96.1832 | rhinopithecus_roxellana | ENSRROP00000026843 | 97.4555 | 96.1832 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 83.4606 | 76.5903 | mus_pahari | MGP_PahariEiJ_P0034306 | 78.8462 | 72.3558 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 91.3486 | 88.5496 | propithecus_coquereli | ENSPCOP00000031758 | 93.2467 | 90.3896 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 38.6768 | 22.1374 | drosophila_melanogaster | FBpp0083753 | 30.7071 | 17.5758 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 48.855 | 32.0611 | ciona_intestinalis | ENSCINP00000003956 | 39.2638 | 25.7669 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 44.5293 | 29.771 | ciona_intestinalis | ENSCINP00000004447 | 41.7661 | 27.9236 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 18.0662 | 8.90585 | eptatretus_burgeri | ENSEBUP00000017237 | 38.7978 | 19.1257 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 62.8499 | 49.3639 | latimeria_chalumnae | ENSLACP00000015915 | 61.596 | 48.3791 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 61.5776 | 48.0916 | lepisosteus_oculatus | ENSLOCP00000017063 | 61.2658 | 47.8481 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 54.4529 | 43.257 | clupea_harengus | ENSCHAP00000039408 | 57.9946 | 46.0705 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 41.4758 | 32.3155 | clupea_harengus | ENSCHAP00000039357 | 51.746 | 40.3175 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 55.7252 | 43.0025 | clupea_harengus | ENSCHAP00000042143 | 55.303 | 42.6768 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 58.7786 | 47.5827 | danio_rerio | ENSDARP00000116736 | 62.6016 | 50.6775 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 54.9618 | 44.2748 | carassius_auratus | ENSCARP00000086785 | 60.6742 | 48.8764 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 56.4885 | 45.5471 | carassius_auratus | ENSCARP00000131382 | 57.2165 | 46.134 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 56.9975 | 43.257 | astyanax_mexicanus | ENSAMXP00000035025 | 56.8528 | 43.1472 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 53.4351 | 43.0025 | astyanax_mexicanus | ENSAMXP00000021366 | 58.9888 | 47.4719 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 59.7964 | 47.8372 | ictalurus_punctatus | ENSIPUP00000031919 | 62.5 | 50 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 56.2341 | 42.7481 | ictalurus_punctatus | ENSIPUP00000028967 | 56.0914 | 42.6396 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 58.5242 | 43.7659 | esox_lucius | ENSELUP00000014319 | 58.9744 | 44.1026 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 61.0687 | 49.3639 | esox_lucius | ENSELUP00000060555 | 53.5714 | 43.3036 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 52.4173 | 37.4046 | electrophorus_electricus | ENSEEEP00000042667 | 56.2842 | 40.1639 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 59.542 | 47.3282 | electrophorus_electricus | ENSEEEP00000033982 | 63.2432 | 50.2703 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 61.3232 | 48.855 | oncorhynchus_kisutch | ENSOKIP00005099681 | 60.8586 | 48.4848 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 57.7608 | 43.7659 | oncorhynchus_kisutch | ENSOKIP00005066943 | 57.9082 | 43.8776 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 58.7786 | 44.0204 | oncorhynchus_kisutch | ENSOKIP00005023724 | 58.1864 | 43.5768 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 57.7608 | 43.7659 | oncorhynchus_mykiss | ENSOMYP00000067785 | 57.0352 | 43.2161 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 59.0331 | 44.2748 | oncorhynchus_mykiss | ENSOMYP00000067749 | 56.3107 | 42.233 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 59.542 | 47.3282 | oncorhynchus_mykiss | ENSOMYP00000051729 | 57.2127 | 45.4768 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 61.0687 | 48.3461 | salmo_salar | ENSSSAP00000057958 | 61.6967 | 48.8432 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 58.0153 | 44.0204 | salmo_salar | ENSSSAP00000161067 | 58.1633 | 44.1327 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 58.0153 | 43.7659 | salmo_trutta | ENSSTUP00000106392 | 58.1633 | 43.8776 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 58.2697 | 43.5115 | salmo_trutta | ENSSTUP00000070576 | 57.25 | 42.75 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 61.5776 | 48.6005 | salmo_trutta | ENSSTUP00000101309 | 61.1111 | 48.2323 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 52.6718 | 39.9491 | gadus_morhua | ENSGMOP00000048877 | 47.2603 | 35.8447 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 52.1628 | 40.9669 | fundulus_heteroclitus | ENSFHEP00000001063 | 58.2386 | 45.7386 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 51.3995 | 40.7125 | poecilia_reticulata | ENSPREP00000019676 | 59.0643 | 46.7836 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 33.8422 | 23.4097 | xiphophorus_maculatus | ENSXMAP00000026894 | 38.8889 | 26.9006 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 52.4173 | 41.7303 | xiphophorus_maculatus | ENSXMAP00000028829 | 60.2339 | 47.9532 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 50.6361 | 38.6768 | oryzias_latipes | ENSORLP00000008031 | 56.5341 | 43.1818 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 55.2163 | 42.4936 | cyclopterus_lumpus | ENSCLMP00005030899 | 56.3636 | 43.3766 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 54.1985 | 40.9669 | oreochromis_niloticus | ENSONIP00000041809 | 56.8 | 42.9333 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 57.2519 | 43.5115 | haplochromis_burtoni | ENSHBUP00000022213 | 57.5448 | 43.734 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 55.4707 | 41.9847 | astatotilapia_calliptera | ENSACLP00000007040 | 56.7708 | 42.9688 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 55.9796 | 43.0025 | sparus_aurata | ENSSAUP00010029879 | 58.0475 | 44.591 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 56.4885 | 42.4936 | lates_calcarifer | ENSLCAP00010016052 | 58.4211 | 43.9474 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 56.9975 | 46.5649 | xenopus_tropicalis | ENSXETP00000053759 | 61.8785 | 50.5525 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 58.2697 | 47.8372 | chrysemys_picta_bellii | ENSCPBP00000031582 | 56.8238 | 46.6501 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 51.6539 | 40.7125 | sphenodon_punctatus | ENSSPUP00000018842 | 64.0379 | 50.4732 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 49.6183 | 39.4402 | laticauda_laticaudata | ENSLLTP00000024105 | 47.1014 | 37.4396 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 46.8193 | 36.8957 | notechis_scutatus | ENSNSUP00000003328 | 48.0418 | 37.859 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 25.1908 | 21.374 | pseudonaja_textilis | ENSPTXP00000020660 | 66.443 | 56.3758 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 34.0967 | 29.2621 | gallus_gallus | ENSGALP00010000467 | 68.7179 | 58.9744 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 23.4097 | 17.5573 | anolis_carolinensis | ENSACAP00000021196 | 60.5263 | 45.3947 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 46.8193 | 35.1145 | neolamprologus_brichardi | ENSNBRP00000026855 | 51.5406 | 38.6555 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 24.173 | 20.3562 | naja_naja | ENSNNAP00000013255 | 61.6883 | 51.9481 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 55.2163 | 41.4758 | podarcis_muralis | ENSPMRP00000017811 | 55.2163 | 41.4758 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 46.3104 | 38.6768 | taeniopygia_guttata | ENSTGUP00000021519 | 54.6547 | 45.6456 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 46.056 | 36.1323 | taeniopygia_guttata | ENSTGUP00000032655 | 42.891 | 33.6493 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 61.3232 | 48.3461 | erpetoichthys_calabaricus | ENSECRP00000015713 | 57.9327 | 45.6731 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 53.4351 | 42.4936 | kryptolebias_marmoratus | ENSKMAP00000018969 | 56.4516 | 44.8925 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 56.2341 | 45.2926 | salvator_merianae | ENSSMRP00000003871 | 56.2341 | 45.2926 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 52.9262 | 40.7125 | oryzias_javanicus | ENSOJAP00000031266 | 59.7701 | 45.977 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 53.6896 | 40.9669 | amphilophus_citrinellus | ENSACIP00000020373 | 59.9432 | 45.7386 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 53.944 | 41.4758 | dicentrarchus_labrax | ENSDLAP00005018261 | 58.8889 | 45.2778 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 55.7252 | 41.7303 | amphiprion_percula | ENSAPEP00000017927 | 58.4 | 43.7333 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 51.6539 | 39.1857 | oryzias_sinensis | ENSOSIP00000001466 | 57.1831 | 43.3803 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 57.7608 | 47.5827 | pelodiscus_sinensis | ENSPSIP00000014905 | 58.2051 | 47.9487 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 57.5064 | 46.5649 | gopherus_evgoodei | ENSGEVP00005020111 | 55.5283 | 44.9631 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 57.2519 | 46.5649 | chelonoidis_abingdonii | ENSCABP00000018274 | 55.2826 | 44.9631 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 57.2519 | 42.2392 | seriola_dumerili | ENSSDUP00000028226 | 59.6817 | 44.0318 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 55.4707 | 42.7481 | cottoperca_gobio | ENSCGOP00000014430 | 58.445 | 45.0402 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 54.1985 | 39.9491 | gasterosteus_aculeatus | ENSGACP00000025614 | 58.1967 | 42.8962 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 53.4351 | 40.2036 | cynoglossus_semilaevis | ENSCSEP00000003997 | 55.7029 | 41.9098 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 51.9084 | 41.2214 | poecilia_formosa | ENSPFOP00000007556 | 59.6491 | 47.3684 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 57.2519 | 41.9847 | myripristis_murdjan | ENSMMDP00005051289 | 59.6817 | 43.7666 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 56.2341 | 42.7481 | sander_lucioperca | ENSSLUP00000039027 | 55.9494 | 42.5316 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 50.8906 | 40.2036 | strigops_habroptila | ENSSHBP00005007544 | 61.7284 | 48.7654 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 55.7252 | 41.7303 | amphiprion_ocellaris | ENSAOCP00000001385 | 58.4 | 43.7333 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 59.0331 | 48.855 | terrapene_carolina_triunguis | ENSTMTP00000028378 | 57.5682 | 47.6427 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 61.8321 | 49.3639 | hucho_hucho | ENSHHUP00000013468 | 56.5116 | 45.1163 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 58.7786 | 44.0204 | hucho_hucho | ENSHHUP00000061550 | 59.6899 | 44.7028 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 58.0153 | 43.5115 | hucho_hucho | ENSHHUP00000072941 | 58.7629 | 44.0722 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 36.6412 | 28.2443 | betta_splendens | ENSBSLP00000045844 | 64.8649 | 50 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 26.7176 | 22.6463 | betta_splendens | ENSBSLP00000034367 | 62.8743 | 53.2934 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 36.8957 | 28.2443 | betta_splendens | ENSBSLP00000035776 | 33.1808 | 25.4005 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 33.0789 | 26.4631 | betta_splendens | ENSBSLP00000048205 | 47.2727 | 37.8182 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 35.1145 | 26.972 | betta_splendens | ENSBSLP00000023147 | 30.5987 | 23.5033 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 34.8601 | 27.4809 | betta_splendens | ENSBSLP00000048405 | 52.8958 | 41.6988 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 25.6997 | 20.8651 | betta_splendens | ENSBSLP00000024075 | 74.2647 | 60.2941 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 37.1501 | 28.4987 | betta_splendens | ENSBSLP00000023807 | 65.4709 | 50.2242 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 35.6234 | 27.4809 | betta_splendens | ENSBSLP00000052325 | 31.0421 | 23.9468 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 26.7176 | 22.6463 | betta_splendens | ENSBSLP00000010759 | 62.8743 | 53.2934 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 36.1323 | 27.9898 | betta_splendens | ENSBSLP00000010864 | 31.4856 | 24.3902 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 27.2265 | 21.6285 | betta_splendens | ENSBSLP00000023746 | 55.7292 | 44.2708 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 37.1501 | 28.4987 | betta_splendens | ENSBSLP00000004751 | 65.4709 | 50.2242 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 33.5878 | 25.6997 | betta_splendens | ENSBSLP00000036367 | 65.6716 | 50.2488 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 31.5522 | 25.1908 | betta_splendens | ENSBSLP00000013302 | 47.6923 | 38.0769 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 37.1501 | 28.7532 | betta_splendens | ENSBSLP00000031009 | 66.6667 | 51.5982 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 27.9898 | 23.1552 | betta_splendens | ENSBSLP00000035494 | 63.2184 | 52.2989 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 34.8601 | 26.972 | betta_splendens | ENSBSLP00000020792 | 63.4259 | 49.0741 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 33.5878 | 24.173 | betta_splendens | ENSBSLP00000023528 | 54.321 | 39.0947 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 37.659 | 28.2443 | betta_splendens | ENSBSLP00000004635 | 36.3636 | 27.2727 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 34.8601 | 26.2087 | betta_splendens | ENSBSLP00000005019 | 55.6911 | 41.8699 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 45.8015 | 34.3511 | betta_splendens | ENSBSLP00000016530 | 53.5714 | 40.1786 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 39.6947 | 30.7888 | betta_splendens | ENSBSLP00000032245 | 53.7931 | 41.7241 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 28.7532 | 23.1552 | betta_splendens | ENSBSLP00000053245 | 58.8542 | 47.3958 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 50.3817 | 38.9313 | betta_splendens | ENSBSLP00000019049 | 55.1532 | 42.6184 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 56.9975 | 44.2748 | pygocentrus_nattereri | ENSPNAP00000014501 | 55.7214 | 43.2836 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 56.2341 | 41.4758 | pygocentrus_nattereri | ENSPNAP00000030579 | 56.8123 | 41.9023 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 55.9796 | 42.7481 | anabas_testudineus | ENSATEP00000028328 | 57.7428 | 44.0945 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 47.8372 | 30.7888 | ciona_savignyi | ENSCSAVP00000016940 | 39.0852 | 25.1559 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 41.4758 | 27.9898 | ciona_savignyi | ENSCSAVP00000016659 | 41.0579 | 27.7078 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 56.9975 | 42.2392 | seriola_lalandi_dorsalis | ENSSLDP00000013209 | 59.4164 | 44.0318 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 55.2163 | 43.0025 | labrus_bergylta | ENSLBEP00000036857 | 58.0214 | 45.1872 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 47.3282 | 33.8422 | pundamilia_nyererei | ENSPNYP00000008816 | 52.1008 | 37.2549 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 55.4707 | 42.2392 | mastacembelus_armatus | ENSMAMP00000005315 | 57.2178 | 43.5696 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 55.4707 | 41.9847 | stegastes_partitus | ENSSPAP00000022019 | 57.2178 | 43.3071 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 61.5776 | 48.855 | oncorhynchus_tshawytscha | ENSOTSP00005027925 | 53.304 | 42.2907 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 59.2875 | 44.7837 | oncorhynchus_tshawytscha | ENSOTSP00005105094 | 58.6902 | 44.3325 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 58.2697 | 43.5115 | oncorhynchus_tshawytscha | ENSOTSP00005013824 | 53.63 | 40.0468 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 55.7252 | 41.9847 | acanthochromis_polyacanthus | ENSAPOP00000027715 | 58.7131 | 44.2359 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 29.5165 | 25.6997 | aquila_chrysaetos_chrysaetos | ENSACCP00020024610 | 55.2381 | 48.0952 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 50.6361 | 37.9135 | nothobranchius_furzeri | ENSNFUP00015003720 | 53.4946 | 40.0538 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 50.3817 | 39.9491 | cyprinodon_variegatus | ENSCVAP00000022398 | 56.4103 | 44.7293 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 51.3995 | 39.1857 | denticeps_clupeoides | ENSDCDP00000033110 | 54.1555 | 41.2869 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 58.5242 | 48.0916 | denticeps_clupeoides | ENSDCDP00000007017 | 59.7403 | 49.0909 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 54.9618 | 42.4936 | larimichthys_crocea | ENSLCRP00005011976 | 60.3352 | 46.648 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 53.1807 | 41.7303 | takifugu_rubripes | ENSTRUP00000030644 | 53.4527 | 41.9437 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 53.1807 | 38.6768 | hippocampus_comes | ENSHCOP00000022970 | 55.5851 | 40.4255 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 54.4529 | 40.9669 | cyprinus_carpio_carpio | ENSCCRP00000062268 | 57.2192 | 43.0481 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 58.5242 | 46.3104 | cyprinus_carpio_carpio | ENSCCRP00000001980 | 53.7383 | 42.5234 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 59.7964 | 47.5827 | cyprinus_carpio_carpio | ENSCCRP00000140175 | 61.1979 | 48.6979 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 51.6539 | 40.9669 | poecilia_latipinna | ENSPLAP00000022694 | 59.3567 | 47.076 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 60.5598 | 47.3282 | sinocyclocheilus_grahami | ENSSGRP00000074183 | 61.4987 | 48.062 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 59.542 | 48.0916 | sinocyclocheilus_grahami | ENSSGRP00000059520 | 61.4173 | 49.6063 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 48.855 | 37.9135 | maylandia_zebra | ENSMZEP00005029097 | 53.4819 | 41.5042 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 53.6896 | 40.9669 | tetraodon_nigroviridis | ENSTNIP00000013035 | 57.967 | 44.2308 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 58.7786 | 49.1094 | paramormyrops_kingsleyae | ENSPKIP00000014805 | 53.5963 | 44.7796 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 53.1807 | 41.2214 | paramormyrops_kingsleyae | ENSPKIP00000021022 | 47.3923 | 36.7347 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 54.1985 | 40.2036 | scleropages_formosus | ENSSFOP00015000080 | 53.25 | 39.5 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 62.0865 | 49.6183 | scleropages_formosus | ENSSFOP00015035795 | 52.8139 | 42.2078 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 54.4529 | 44.2748 | leptobrachium_leishanense | ENSLLEP00000038025 | 56.7639 | 46.1538 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 56.9975 | 43.257 | scophthalmus_maximus | ENSSMAP00000015064 | 58.7927 | 44.6194 |
| ENSG00000141510 | homo_sapiens | ENSP00000269305 | 52.1628 | 40.458 | oryzias_melastigma | ENSOMEP00000009309 | 59.2486 | 45.9538 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0000122 | involved_in negative regulation of transcription by RNA polymerase II | 2 | IDA,ISS | Homo_sapiens(9606) | Process |
| GO:0000423 | involved_in mitophagy | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0000785 | part_of chromatin | 3 | IDA,IMP,ISA | Homo_sapiens(9606) | Component |
| GO:0000976 | enables transcription cis-regulatory region binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0000978 | enables RNA polymerase II cis-regulatory region sequence-specific DNA binding | 3 | IBA,IDA,ISS | Homo_sapiens(9606) | Function |
| GO:0000981 | enables DNA-binding transcription factor activity, RNA polymerase II-specific | 3 | IBA,IDA,ISA | Homo_sapiens(9606) | Function |
| GO:0000987 | enables cis-regulatory region sequence-specific DNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0001046 | enables core promoter sequence-specific DNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0001094 | enables TFIID-class transcription factor complex binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0001227 | enables DNA-binding transcription repressor activity, RNA polymerase II-specific | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0001228 | enables DNA-binding transcription activator activity, RNA polymerase II-specific | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0001701 | involved_in in utero embryonic development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0001756 | involved_in somitogenesis | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0001836 | involved_in release of cytochrome c from mitochondria | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0002020 | enables protease binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0002039 | enables p53 binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0002244 | involved_in hematopoietic progenitor cell differentiation | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0002309 | involved_in T cell proliferation involved in immune response | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0002326 | involved_in B cell lineage commitment | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0002360 | involved_in T cell lineage commitment | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0002931 | involved_in response to ischemia | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0003677 | enables DNA binding | 2 | IDA,IMP | Homo_sapiens(9606) | Function |
| GO:0003682 | enables chromatin binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0003700 | enables DNA-binding transcription factor activity | 2 | IDA,IMP | Homo_sapiens(9606) | Function |
| GO:0003730 | enables mRNA 3-UTR binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0005507 | enables copper ion binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005634 | located_in nucleus | 2 | IDA,IMP | Homo_sapiens(9606) | Component |
| GO:0005654 | located_in nucleoplasm | 2 | IDA,TAS | Homo_sapiens(9606) | Component |
| GO:0005657 | located_in replication fork | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0005667 | part_of transcription regulator complex | 1 | IGI | Homo_sapiens(9606) | Component |
| GO:0005730 | located_in nucleolus | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005737 | located_in cytoplasm | 2 | IDA,IMP | Homo_sapiens(9606) | Component |
| GO:0005739 | located_in mitochondrion | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005759 | located_in mitochondrial matrix | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0005783 | located_in endoplasmic reticulum | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0005813 | located_in centrosome | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005829 | located_in cytosol | 2 | IDA,TAS | Homo_sapiens(9606) | Component |
| GO:0006289 | involved_in nucleotide-excision repair | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0006302 | involved_in double-strand break repair | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0006355 | involved_in regulation of DNA-templated transcription | 2 | IDA,IMP | Homo_sapiens(9606) | Process |
| GO:0006357 | involved_in regulation of transcription by RNA polymerase II | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0006606 | involved_in protein import into nucleus | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0006914 | involved_in autophagy | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0006974 | acts_upstream_of_or_within DNA damage response | 3 | IDA,IMP | Homo_sapiens(9606) | Process |
| GO:0006977 | involved_in DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0006978 | involved_in DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator | 2 | IDA,IMP | Homo_sapiens(9606) | Process |
| GO:0006983 | acts_upstream_of_or_within ER overload response | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0007179 | involved_in transforming growth factor beta receptor signaling pathway | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0007265 | involved_in Ras protein signal transduction | 1 | IEP | Homo_sapiens(9606) | Process |
| GO:0007369 | involved_in gastrulation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0007405 | involved_in neuroblast proliferation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0007406 | involved_in negative regulation of neuroblast proliferation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0008104 | involved_in protein localization | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0008156 | involved_in negative regulation of DNA replication | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0008270 | enables zinc ion binding | 1 | TAS | Homo_sapiens(9606) | Function |
| GO:0008285 | involved_in negative regulation of cell population proliferation | 4 | IDA,IMP,ISS | Homo_sapiens(9606) | Process |
| GO:0008340 | involved_in determination of adult lifespan | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0009299 | involved_in mRNA transcription | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0009303 | involved_in rRNA transcription | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0009651 | involved_in response to salt stress | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0010035 | involved_in response to inorganic substance | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0010165 | involved_in response to X-ray | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0010332 | involved_in response to gamma radiation | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0010628 | involved_in positive regulation of gene expression | 4 | IDA,IMP,ISS | Homo_sapiens(9606) | Process |
| GO:0010659 | involved_in cardiac muscle cell apoptotic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0010666 | involved_in positive regulation of cardiac muscle cell apoptotic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0014009 | involved_in glial cell proliferation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0016032 | involved_in viral process | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0016363 | located_in nuclear matrix | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0016604 | colocalizes_with nuclear body | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0016605 | located_in PML body | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0017053 | part_of transcription repressor complex | 1 | IPI | Homo_sapiens(9606) | Component |
| GO:0019661 | involved_in glucose catabolic process to lactate via pyruvate | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0019899 | enables enzyme binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0021549 | involved_in cerebellum development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0030308 | involved_in negative regulation of cell growth | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0030330 | involved_in DNA damage response, signal transduction by p53 class mediator | 2 | IDA,IMP | Homo_sapiens(9606) | Process |
| GO:0030512 | involved_in negative regulation of transforming growth factor beta receptor signaling pathway | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0030971 | enables receptor tyrosine kinase binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0031571 | involved_in mitotic G1 DNA damage checkpoint signaling | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0031625 | enables ubiquitin protein ligase binding | 2 | EXP,IPI | Homo_sapiens(9606) | Function |
| GO:0032991 | part_of protein-containing complex | 2 | IDA,IMP | Homo_sapiens(9606) | Component |
| GO:0033077 | involved_in T cell differentiation in thymus | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0033209 | involved_in tumor necrosis factor-mediated signaling pathway | 1 | IGI | Homo_sapiens(9606) | Process |
| GO:0034103 | involved_in regulation of tissue remodeling | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0034644 | involved_in cellular response to UV | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0035033 | enables histone deacetylase regulator activity | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0035264 | involved_in multicellular organism growth | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0035794 | involved_in positive regulation of mitochondrial membrane permeability | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0035861 | located_in site of double-strand break | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0036003 | involved_in positive regulation of transcription from RNA polymerase II promoter in response to stress | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0036310 | enables ATP-dependent DNA/DNA annealing activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0042149 | acts_upstream_of_or_within cellular response to glucose starvation | 2 | IDA | Homo_sapiens(9606) | Process |
| GO:0042771 | acts_upstream_of_or_within intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 3 | IDA,IMP | Homo_sapiens(9606) | Process |
| GO:0042802 | enables identical protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0042826 | enables histone deacetylase binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0042981 | acts_upstream_of_or_within regulation of apoptotic process | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0043065 | involved_in positive regulation of apoptotic process | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0043066 | involved_in negative regulation of apoptotic process | 2 | IGI,IMP | Homo_sapiens(9606) | Process |
| GO:0043073 | located_in germ cell nucleus | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0043153 | involved_in entrainment of circadian clock by photoperiod | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0043504 | involved_in mitochondrial DNA repair | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0043516 | involved_in regulation of DNA damage response, signal transduction by p53 class mediator | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0043525 | involved_in positive regulation of neuron apoptotic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0043621 | enables protein self-association | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0045815 | involved_in transcription initiation-coupled chromatin remodeling | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0045861 | involved_in negative regulation of proteolysis | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0045892 | involved_in negative regulation of DNA-templated transcription | 3 | IDA,IMP,NAS | Homo_sapiens(9606) | Process |
| GO:0045893 | involved_in positive regulation of DNA-templated transcription | 2 | IDA,IMP | Homo_sapiens(9606) | Process |
| GO:0045899 | involved_in positive regulation of RNA polymerase II transcription preinitiation complex assembly | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0045944 | acts_upstream_of_or_within positive regulation of transcription by RNA polymerase II | 4 | IDA,IGI,IMP | Homo_sapiens(9606) | Process |
| GO:0046677 | involved_in response to antibiotic | 1 | IEP | Homo_sapiens(9606) | Process |
| GO:0046982 | enables protein heterodimerization activity | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0048144 | involved_in fibroblast proliferation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0048147 | involved_in negative regulation of fibroblast proliferation | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0048512 | involved_in circadian behavior | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0048539 | involved_in bone marrow development | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0048568 | involved_in embryonic organ development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0050731 | involved_in positive regulation of peptidyl-tyrosine phosphorylation | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0050821 | involved_in protein stabilization | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0051087 | enables protein-folding chaperone binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0051097 | involved_in negative regulation of helicase activity | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0051262 | involved_in protein tetramerization | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0051276 | involved_in chromosome organization | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0051402 | involved_in neuron apoptotic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0051721 | enables protein phosphatase 2A binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0051726 | involved_in regulation of cell cycle | 4 | IDA,IGI,IMP,ISS | Homo_sapiens(9606) | Process |
| GO:0051974 | involved_in negative regulation of telomerase activity | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0060218 | involved_in hematopoietic stem cell differentiation | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0060253 | involved_in negative regulation of glial cell proliferation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0060333 | involved_in type II interferon-mediated signaling pathway | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0060411 | involved_in cardiac septum morphogenesis | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0061419 | involved_in positive regulation of transcription from RNA polymerase II promoter in response to hypoxia | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0061629 | enables RNA polymerase II-specific DNA-binding transcription factor binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0062100 | involved_in positive regulation of programmed necrotic cell death | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0065003 | involved_in protein-containing complex assembly | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0070059 | involved_in intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0070242 | involved_in thymocyte apoptotic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0070245 | involved_in positive regulation of thymocyte apoptotic process | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0070266 | involved_in necroptotic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0071456 | involved_in cellular response to hypoxia | 1 | IEP | Homo_sapiens(9606) | Process |
| GO:0071466 | involved_in cellular response to xenobiotic stimulus | 1 | IEP | Homo_sapiens(9606) | Process |
| GO:0071479 | involved_in cellular response to ionizing radiation | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0071480 | involved_in cellular response to gamma radiation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0071494 | involved_in cellular response to UV-C | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0071889 | enables 14-3-3 protein binding | 1 | EXP | Homo_sapiens(9606) | Function |
| GO:0072089 | involved_in stem cell proliferation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0072331 | involved_in signal transduction by p53 class mediator | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0072332 | involved_in intrinsic apoptotic signaling pathway by p53 class mediator | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0072593 | involved_in reactive oxygen species metabolic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0072717 | involved_in cellular response to actinomycin D | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0090200 | involved_in positive regulation of release of cytochrome c from mitochondria | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0090398 | involved_in cellular senescence | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0090399 | involved_in replicative senescence | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0090403 | involved_in oxidative stress-induced premature senescence | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0097193 | involved_in intrinsic apoptotic signaling pathway | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0097252 | involved_in oligodendrocyte apoptotic process | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0097371 | enables MDM2/MDM4 family protein binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0097718 | enables disordered domain specific binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0140296 | enables general transcription initiation factor binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0140677 | enables molecular function activator activity | 2 | EXP,IPI | Homo_sapiens(9606) | Function |
| GO:1900119 | involved_in positive regulation of execution phase of apoptosis | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:1901525 | involved_in negative regulation of mitophagy | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:1902108 | involved_in regulation of mitochondrial membrane permeability involved in apoptotic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:1902253 | involved_in regulation of intrinsic apoptotic signaling pathway by p53 class mediator | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:1902749 | acts_upstream_of_negative_effect regulation of cell cycle G2/M phase transition | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:1902895 | involved_in positive regulation of miRNA transcription | 3 | IDA,IGI,IMP | Homo_sapiens(9606) | Process |
| GO:1903451 | involved_in negative regulation of G1 to G0 transition | 2 | IDA,IMP | Homo_sapiens(9606) | Process |
| GO:1903799 | involved_in negative regulation of miRNA processing | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:1904024 | involved_in negative regulation of glucose catabolic process to lactate via pyruvate | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:1905856 | involved_in negative regulation of pentose-phosphate shunt | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:1990144 | involved_in intrinsic apoptotic signaling pathway in response to hypoxia | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:1990440 | involved_in positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:1990841 | enables promoter-specific chromatin binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:2000269 | involved_in regulation of fibroblast apoptotic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:2000378 | involved_in negative regulation of reactive oxygen species metabolic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:2000379 | involved_in positive regulation of reactive oxygen species metabolic process | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:2000647 | involved_in negative regulation of stem cell proliferation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:2000774 | involved_in positive regulation of cellular senescence | 2 | IDA,ISS | Homo_sapiens(9606) | Process |
| GO:2001244 | involved_in positive regulation of intrinsic apoptotic signaling pathway | 2 | IMP,ISS | Homo_sapiens(9606) | Process |