Welcome to
RBP World!
Functions and Diseases
| RBP Type | Canonical_RBPs |
| Diseases | Cancer Disease |
| Drug | Drug |
| Main interacting RNAs | mRNA |
| Moonlighting functions | Transmemebrane |
| Localizations | N.A. |
| BulkPerturb-seq | DataSet_01_186 DataSet_01_276 |
Description
| Ensembl ID | ENSG00000140718 | Gene ID | 79068 | Accession | 24678 |
| Symbol | FTO | Alias | GDFD;IFEX9;ALKBH9;BMIQ14 | Full Name | FTO alpha-ketoglutarate dependent dioxygenase |
| Status | Confidence | Length | 456821 bases | Strand | Plus strand |
| Position | 16 : 53701692 - 54158512 | RNA binding domain | FTO_NTD , FTO_CTD | ||
| Summary | This gene is a nuclear protein of the AlkB related non-haem iron and 2-oxoglutarate-dependent oxygenase superfamily but the exact physiological function of this gene is not known. Other non-heme iron enzymes function to reverse alkylated DNA and RNA damage by oxidative demethylation. Studies in mice and humans indicate a role in nervous and cardiovascular systems and a strong association with body mass index, obesity risk, and type 2 diabetes. [provided by RefSeq, Jul 2011] | ||||
RNA binding domains (RBDs)
| Protein | Domain | Pfam ID | E-value | Domain number |
|---|---|---|---|---|
| ENSP00000490300 | FTO_CTD | PF12934 | 1.9e-58 | 1 |
| ENSP00000418823 | FTO_CTD | PF12934 | 2.1e-57 | 1 |
| ENSP00000418823 | FTO_NTD | PF12933 | 1.4e-126 | 2 |
| ENSP00000489638 | FTO_CTD | PF12934 | 3.9e-57 | 1 |
| ENSP00000489638 | FTO_NTD | PF12933 | 1.6e-126 | 2 |
| ENSP00000490516 | FTO_CTD | PF12934 | 3.9e-57 | 1 |
| ENSP00000490516 | FTO_NTD | PF12933 | 1.9e-126 | 2 |
| ENSP00000489936 | FTO_CTD | PF12934 | 7.9e-43 | 1 |
| ENSP00000489936 | FTO_NTD | PF12933 | 1e-126 | 2 |
| ENSP00000489714 | FTO_CTD | PF12934 | 8e-42 | 1 |
| ENSP00000489714 | FTO_NTD | PF12933 | 1.3e-62 | 2 |
| ENSP00000489641 | FTO_CTD | PF12934 | 1.6e-41 | 1 |
| ENSP00000489641 | FTO_NTD | PF12933 | 1.1e-126 | 2 |
| ENSP00000490047 | FTO_CTD | PF12934 | 6.7e-41 | 1 |
| ENSP00000490047 | FTO_NTD | PF12933 | 1.1e-126 | 2 |
| ENSP00000490426 | FTO_CTD | PF12934 | 9e-41 | 1 |
| ENSP00000490426 | FTO_NTD | PF12933 | 1.1e-126 | 2 |
| ENSP00000489886 | FTO_CTD | PF12934 | 2e-30 | 1 |
| ENSP00000489886 | FTO_NTD | PF12933 | 8.9e-127 | 2 |
| ENSP00000415636 | FTO_CTD | PF12934 | 5e-26 | 1 |
| ENSP00000417422 | FTO_CTD | PF12934 | 5e-26 | 1 |
| ENSP00000417843 | FTO_CTD | PF12934 | 7.9e-21 | 1 |
| ENSP00000489904 | FTO_CTD | PF12934 | 7.8e-14 | 1 |
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|---|---|---|---|
| 36814457 | A computational map of the human-SARS-CoV-2 protein-RNA interactome predicted at single-nucleotide resolution. | Marc Horlacher | 2023-03-01 | NAR genomics and bioinformatics |
| 35605980 | Enzymatic Characterization of In Vitro Activity of RNA Methyltransferase PCIF1 on DNA. | Dan Yu | 2022-05-23 | Biochemistry |
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| FTO-215 | ENST00000636491 | 1676 | 450aa | ENSP00000490047 |
| FTO-214 | ENST00000636218 | 2227 | 457aa | ENSP00000489641 |
| FTO-217 | ENST00000637001 | 1609 | 481aa | ENSP00000489936 |
| FTO-209 | ENST00000570395 | 555 | No protein | - |
| FTO-216 | ENST00000636992 | 1514 | 421aa | ENSP00000489886 |
| FTO-206 | ENST00000471389 | 11573 | 505aa | ENSP00000418823 |
| FTO-222 | ENST00000637969 | 2889 | 559aa | ENSP00000490516 |
| FTO-212 | ENST00000636030 | 1636 | No protein | - |
| FTO-220 | ENST00000637562 | 1524 | 455aa | ENSP00000490426 |
| FTO-221 | ENST00000637845 | 2472 | 533aa | ENSP00000489638 |
| FTO-205 | ENST00000464071 | 1065 | 70aa | ENSP00000418424 |
| FTO-208 | ENST00000563011 | 2015 | 275aa | ENSP00000489714 |
| FTO-210 | ENST00000612285 | 1852 | 173aa | ENSP00000490300 |
| FTO-202 | ENST00000431610 | 707 | 106aa | ENSP00000415636 |
| FTO-203 | ENST00000460382 | 1517 | 106aa | ENSP00000417422 |
| FTO-219 | ENST00000637062 | 459 | 66aa | ENSP00000489904 |
| FTO-204 | ENST00000463855 | 3056 | 127aa | ENSP00000417843 |
| FTO-211 | ENST00000635892 | 1085 | No protein | - |
| FTO-201 | ENST00000268349 | 2799 | 129aa | ENSP00000268349 |
| FTO-207 | ENST00000472835 | 847 | No protein | - |
| FTO-213 | ENST00000636091 | 5323 | No protein | - |
| FTO-218 | ENST00000637049 | 2105 | 61aa | ENSP00000489891 |
Phenotypes
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| 17828 | Body Mass Index | 2.237e-005 | 17658951 |
| 17831 | Body Mass Index | 3.378e-005 | 17658951 |
| 17838 | Body Mass Index | 1.802e-005 | 17658951 |
| 17844 | Body Mass Index | 1.802e-005 | 17658951 |
| 17847 | Body Mass Index | 3.954e-005 | 17658951 |
| 17848 | Body Mass Index | 1.013e-005 | 17658951 |
| 17849 | Body Mass Index | 1.802e-005 | 17658951 |
| 17850 | Body Mass Index | 2.767e-005 | 17658951 |
| 17851 | Body Mass Index | 2.604e-005 | 17658951 |
| 17856 | Body Mass Index | 7.241e-005 | 17658951 |
| 17860 | Body Weights and Measures | 9.547e-006 | 17658951 |
| 17864 | Body Weights and Measures | 6.989e-005 | 17658951 |
| 17865 | Body Weights and Measures | 2.954e-005 | 17658951 |
| 17875 | Body Weights and Measures | 3.42e-005 | 17658951 |
| 17880 | Body Weights and Measures | 3.42e-005 | 17658951 |
| 17883 | Body Weights and Measures | 8.726e-005 | 17658951 |
| 17884 | Body Weights and Measures | 1.466e-005 | 17658951 |
| 17885 | Body Weights and Measures | 3.42e-005 | 17658951 |
| 17886 | Body Weights and Measures | 1.052e-005 | 17658951 |
| 17887 | Body Weights and Measures | 1.007e-005 | 17658951 |
| 18070 | Waist Circumference | 3.141e-005 | - |
| 18071 | Waist Circumference | 1.128e-005 | - |
| 18116 | Body Weight | 1.503e-005 | 17658951 |
| 18117 | Body Weight | 4.917e-006 | 17658951 |
| 19049 | Acceleration | 7.888e-005 | - |
| 19512 | Blood Pressure | 9.284e-005 | - |
| 19519 | Blood Pressure | 7.618e-005 | - |
| 27739 | Coronary Disease | 8.6460000E-005 | - |
| 30340 | Potassium | 2.7840000E-005 | - |
| 32679 | Body Mass Index | 3.3947667E-005 | - |
| 32805 | Body Mass Index | 9.5155245E-005 | - |
| 32895 | Body Mass Index | 9.9574260E-005 | - |
| 35508 | Platelet Function Tests | 2.5876800E-005 | - |
| 38988 | Platelet Function Tests | 1.5066100E-005 | - |
| 41344 | Platelet Function Tests | 2.2874400E-005 | - |
| 44578 | Platelet Function Tests | 8.3500973E-006 | - |
| 47709 | Intelligence Tests | 1.6674600E-005 | - |
| 47715 | Intelligence Tests | 2.1792800E-005 | - |
| 48487 | Child Development Disorders, Pervasive | 2.1737300E-005 | - |
| 48489 | Child Development Disorders, Pervasive | 2.8861500E-005 | - |
| 49308 | Child Development Disorders, Pervasive | 3.3854000E-005 | - |
| 50031 | Child Development Disorders, Pervasive | 1.3875000E-006 | - |
| 50035 | Child Development Disorders, Pervasive | 4.0257000E-006 | - |
| 54279 | Adiponectin | 7.0640000E-006 | - |
| 54285 | Adiponectin | 7.4040000E-005 | - |
| 55662 | 25-Hydroxyvitamin D 2 | 3.6780000E-005 | - |
| 60765 | Obesity | 1E-28 | 19151714 |
| 61016 | Obesity | 7E-18 | 19151714 |
| 61038 | Obesity | 5E-13 | 19151714 |
| 61466 | Breast Neoplasms | 6E-7 | 23535733 |
| 62114 | Body Fat Distribution | 1E-13 | 26833246 |
| 62208 | Obesity | 1E-7 | 21544081 |
| 63142 | Body Mass Index | 3E-35 | 17434869 |
| 63430 | Body Mass Index | 5E-120 | 20935630 |
| 63436 | Diabetes Mellitus, Type 2 | 3E-8 | 20581827 |
| 63628 | Obesity | 6E-12 | 19553259 |
| 64654 | Obesity | 5E-110 | 23563607 |
| 65208 | Obesity | 6E-39 | 23563607 |
| 65257 | Hair | 3E-6 | 26926045 |
| 65985 | Body Mass Index | 8E-153 | 25673413 |
| 66380 | Hip | 3E-52 | 25673412 |
| 66566 | Waist-Hip Ratio | 1E-17 | 25673412 |
| 67152 | Body Mass Index | 2E-7 | 19396169 |
| 67365 | Rhinitis, Allergic | 6E-7 | 25085501 |
| 68066 | Breast | 4E-7 | 27182965 |
| 68081 | Subcutaneous Fat | 6E-8 | 22589738 |
| 68181 | Body Mass Index | 4E-84 | 25673413 |
| 68837 | Diabetes Mellitus, Type 2 | 7E-14 | 17463249 |
| 69335 | Diabetes Mellitus, Type 2 | 1E-12 | 17463248 |
| 69370 | Body Mass Index | 2E-10 | 22982992 |
| 69663 | Breast Neoplasms | 3E-7 | 27117709 |
| 69683 | Osteoarthritis | 4E-6 | 22763110 |
| 70456 | Body Mass Index | 1E-7 | 24348519 |
| 70983 | Waist Circumference | 4E-101 | 25673412 |
| 71377 | Body Weight | 6E-14 | 19079260 |
| 71456 | Body Mass Index | 1E-47 | 19079260 |
| 71848 | Diabetes Mellitus, Type 2 | 2E-17 | 19056611 |
| 72021 | Menarche | 2E-17 | 25231870 |
| 72464 | Body Mass Index | 2E-16 | 25953783 |
| 72556 | Obesity | 3E-26 | 21706003 |
| 73592 | Body Mass Index | 1E-93 | 25673413 |
| 74017 | Body Mass Index | 1E-10 | 23583978 |
| 74039 | Melanoma | 4E-12 | 28212542 |
| 75161 | Melanoma | 4E-12 | 23455637 |
| 75334 | Body Mass Index | 4E-14 | 26604143 |
| 75534 | Waist Circumference | 8E-59 | 25673412 |
| 76027 | Menarche | 3E-8 | 21102462 |
| 76766 | Subcutaneous Fat | 3E-6 | 22589738 |
| 76999 | Obesity | 1E-7 | 18159244 |
| 77171 | Obesity | 2E-12 | 21552555 |
| 78199 | Breast Neoplasms | 2E-6 | 27117709 |
| 78270 | Body Mass Index | 2E-51 | 23563607 |
| 78830 | Menarche | 1E-12 | 27182965 |
| 79236 | Body Mass Index | 4E-13 | 19079260 |
| 79731 | Body Weight | 5E-36 | 19079260 |
| 79822 | Waist Circumference | 5E-19 | 19557197 |
| 80088 | Body Fat Distribution | 1E-25 | 26833246 |
| 80542 | Obesity | 1E-79 | 23563607 |
| 81093 | Metabolic Syndrome X | 2E-9 | 22399527 |
| 81873 | Leptin | 1E-9 | 26833098 |
| 82635 | Cholesterol, HDL | 7E-9 | 24097068 |
| 82753 | Hip | 4E-43 | 25673412 |
| 82800 | Atrial Fibrillation | 1E-14 | 27790247 |
| 82801 | Blood Pressure | 1E-14 | 27790247 |
| 82802 | Cholesterol | 1E-14 | 27790247 |
| 82803 | Coronary Disease | 1E-14 | 27790247 |
| 82804 | Diabetes Mellitus | 1E-14 | 27790247 |
| 82805 | Electrocardiography | 1E-14 | 27790247 |
| 82806 | Glucose | 1E-14 | 27790247 |
| 82807 | Heart Failure | 1E-14 | 27790247 |
| 82808 | Hematocrit | 1E-14 | 27790247 |
| 82809 | Cholesterol, HDL | 1E-14 | 27790247 |
| 82810 | Mortality | 1E-14 | 27790247 |
| 82811 | Neoplasms | 1E-14 | 27790247 |
| 82812 | Triglycerides | 1E-14 | 27790247 |
| 82813 | Body Mass Index | 1E-14 | 27790247 |
| 82814 | Stroke | 1E-14 | 27790247 |
| 83609 | Body Mass Index | 7E-27 | 24861553 |
| 83867 | Breast Neoplasms | 3E-8 | 27117709 |
| 85566 | Body Fat Distribution | 4E-27 | 26833246 |
| 85683 | Body Mass Index | 4E-51 | 19079261 |
| 87592 | Diabetes Mellitus, Type 2 | 2E-7 | 17554300 |
| 87745 | Diabetes Mellitus, Type 2 | 1E-20 | 22693455 |
| 87932 | Waist-Hip Ratio | 1E-38 | 25673412 |
| 87974 | Breast Neoplasms | 4E-8 | 23535733 |
| 89025 | Diabetes Mellitus, Type 2 | 7E-6 | 18372903 |
| 89132 | Body Fat Distribution | 7E-14 | 26833246 |
| 89879 | Body Mass Index | 5E-22 | 22344221 |
| 90064 | Body Fat Distribution | 2E-13 | 26833246 |
| 90339 | Body Mass Index | 1E-26 | 25953783 |
| 91444 | Diabetes Mellitus, Type 2 | 1E-12 | 24509480 |
| 91658 | Sex Hormone-Binding Globulin | 9E-6 | 22675492 |
| 92459 | Body Mass Index | 5E-16 | 26604143 |
| 93969 | Obesity | 3E-28 | 23563609 |
| 96067 | Diabetes Mellitus, Type 2 | 6E-6 | 23209189 |
| 96360 | Body Mass Index | 1E-26 | 25953783 |
| 96622 | Body Mass Index | 9E-10 | 25044758 |
| 98784 | Diabetes Mellitus, Type 2 | 4E-15 | 26818947 |
| 98796 | Body Mass Index | 2E-14 | 24064335 |
| 99389 | Leukopenia | 1E-8 | 27558924 |
| 99390 | Purines | 1E-8 | 27558924 |
| 99391 | Inflammatory Bowel Diseases | 1E-8 | 27558924 |
| 99609 | Obesity | 5E-19 | 20421936 |
| 99631 | Obesity | 2E-81 | 23563607 |
| 100591 | Waist-Hip Ratio | 1E-23 | 25673412 |
| 100651 | C-Reactive Protein | 5E-9 | 27286809 |
| 100652 | Cholesterol, HDL | 5E-9 | 27286809 |
| 100756 | Body Mass Index | 4E-23 | 23669352 |
| 101039 | Body Mass Index | 1E-156 | 25673413 |
| 101162 | Energy Intake | 1E-9 | 23636237 |
| 101312 | Body Mass Index | 4E-8 | 21037115 |
| 101313 | Pulmonary Disease, Chronic Obstructive | 4E-8 | 21037115 |
| 102681 | Body Fat Distribution | 4E-15 | 26833246 |
| 102864 | Breast Neoplasms | 5E-8 | 27117709 |
| 102984 | Waist Circumference | 1E-57 | 25673412 |
| 103167 | Leptin | 9E-8 | 26833098 |
| 103468 | Breast Neoplasms | 6E-14 | 23535729 |
| 104257 | Body Mass Index | 4E-8 | 18454148 |
| 104610 | sam so eum | 1E-7 | 25888059 |
| 104611 | Medicine, Korean Traditional | 1E-7 | 25888059 |
| 104669 | Hip | 2E-86 | 25673412 |
| 104820 | Body Weight | 9E-7 | 17658951 |
| 104821 | Waist-Hip Ratio | 3E-8 | 17658951 |
| 104822 | Body Mass Index | 9E-7 | 17658951 |
| 105695 | Blood Pressure | 2E-17 | 27790247 |
| 105696 | Cholesterol | 2E-17 | 27790247 |
| 105697 | Electrocardiography | 2E-17 | 27790247 |
| 105698 | Glucose | 2E-17 | 27790247 |
| 105699 | Hematocrit | 2E-17 | 27790247 |
| 105700 | Cholesterol, HDL | 2E-17 | 27790247 |
| 105701 | Triglycerides | 2E-17 | 27790247 |
| 105702 | Body Mass Index | 2E-17 | 27790247 |
| 106785 | Subcutaneous Fat | 1E-9 | 27918534 |
| 106864 | Triglycerides | 3E-8 | 24097068 |
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
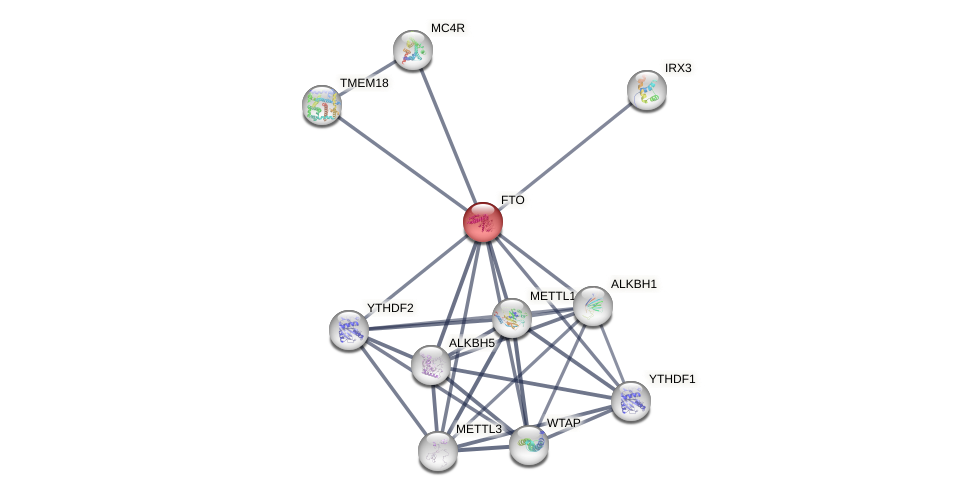
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000140718 | ||||||||
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 98.6139 | 98.2178 | nomascus_leucogenys | ENSNLEP00000041700 | 89.0877 | 88.7299 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 99.0099 | 98.8119 | pan_paniscus | ENSPPAP00000009628 | 89.4454 | 89.2665 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 98.2178 | 97.6238 | pongo_abelii | ENSPPYP00000028640 | 88.7299 | 88.1932 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 98.8119 | 98.6139 | pan_troglodytes | ENSPTRP00000093800 | 89.2665 | 89.0877 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 99.0099 | 99.0099 | gorilla_gorilla | ENSGGOP00000034557 | 89.4454 | 89.4454 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 53.2673 | 44.1584 | ornithorhynchus_anatinus | ENSOANP00000021164 | 68.798 | 57.0332 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 24.3564 | 21.1881 | ornithorhynchus_anatinus | ENSOANP00000038265 | 76.3975 | 66.4596 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 82.3762 | 69.3069 | sarcophilus_harrisii | ENSSHAP00000022477 | 82.8685 | 69.7211 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 73.8614 | 62.9703 | notamacropus_eugenii | ENSMEUP00000010306 | 74.3028 | 63.3466 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 55.8416 | 53.8614 | choloepus_hoffmanni | ENSCHOP00000001108 | 55.8416 | 53.8614 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 39.0099 | 37.6238 | dasypus_novemcinctus | ENSDNOP00000029277 | 94.2584 | 90.9091 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 85.9406 | 78.0198 | echinops_telfairi | ENSETEP00000010148 | 85.9406 | 78.0198 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 98.2178 | 97.0297 | callithrix_jacchus | ENSCJAP00000065416 | 88.7299 | 87.6565 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 99.0099 | 98.4158 | cercocebus_atys | ENSCATP00000013210 | 99.0099 | 98.4158 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 98.0198 | 97.4257 | macaca_fascicularis | ENSMFAP00000020089 | 89.3502 | 88.8087 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 97.8218 | 97.0297 | macaca_mulatta | ENSMMUP00000079401 | 83.165 | 82.4916 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 97.6238 | 96.6337 | macaca_nemestrina | ENSMNEP00000044970 | 86.9489 | 86.067 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 97.4257 | 96.8317 | papio_anubis | ENSPANP00000040946 | 85.2686 | 84.7487 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 98.4158 | 97.8218 | mandrillus_leucophaeus | ENSMLEP00000004039 | 99.004 | 98.4064 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 86.5347 | 82.1782 | tursiops_truncatus | ENSTTRP00000012061 | 86.5347 | 82.1782 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 87.1287 | 82.9703 | vulpes_vulpes | ENSVVUP00000026593 | 76.9231 | 73.2517 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 92.8713 | 86.9307 | mus_spretus | MGP_SPRETEiJ_P0089012 | 93.4263 | 87.4502 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 94.8515 | 91.0891 | equus_asinus | ENSEASP00005054084 | 86.7754 | 83.3333 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 94.4554 | 87.5247 | capra_hircus | ENSCHIP00000004985 | 94.4554 | 87.5247 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 93.2673 | 88.1188 | loxodonta_africana | ENSLAFP00000013117 | 93.2673 | 88.1188 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 94.6535 | 90.8911 | panthera_leo | ENSPLOP00000009404 | 78.4893 | 75.3695 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 19.2079 | 18.8119 | ailuropoda_melanoleuca | ENSAMEP00000035080 | 91.5094 | 89.6226 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 93.0693 | 85.1485 | bos_indicus_hybrid | ENSBIXP00005034074 | 90.2111 | 82.5336 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 95.6436 | 91.8812 | oryctolagus_cuniculus | ENSOCUP00000031800 | 95.8333 | 92.0635 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 94.6535 | 90.6931 | equus_caballus | ENSECAP00000038168 | 89.8496 | 86.0902 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 94.2574 | 90.495 | canis_lupus_familiaris | ENSCAFP00845003351 | 76.7742 | 73.7097 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 94.8515 | 91.0891 | panthera_pardus | ENSPPRP00000016701 | 95.0397 | 91.2698 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 96.2376 | 91.8812 | marmota_marmota_marmota | ENSMMMP00000013135 | 96.4286 | 92.0635 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 95.0495 | 90.495 | balaenoptera_musculus | ENSBMSP00010022501 | 92.4855 | 88.0539 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 92.0792 | 87.5247 | cavia_porcellus | ENSCPOP00000011645 | 93.1864 | 88.5772 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 93.8614 | 90.099 | felis_catus | ENSFCAP00000004882 | 90.9789 | 87.3321 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 91.4851 | 84.7525 | ursus_americanus | ENSUAMP00000015686 | 85.2399 | 78.9668 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 80.5941 | 66.9307 | monodelphis_domestica | ENSMODP00000048198 | 77.9693 | 64.751 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 94.6535 | 87.7228 | bison_bison_bison | ENSBBBP00000017081 | 94.6535 | 87.7228 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 95.2475 | 90.495 | monodon_monoceros | ENSMMNP00015012758 | 95.2475 | 90.495 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 91.8812 | 87.1287 | sus_scrofa | ENSSSCP00000047110 | 91.1591 | 86.444 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 95.6436 | 93.2673 | microcebus_murinus | ENSMICP00000047022 | 92.5287 | 90.2299 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 91.8812 | 87.3267 | octodon_degus | ENSODEP00000005892 | 92.4303 | 87.8486 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 92.2772 | 86.1386 | jaculus_jaculus | ENSJJAP00000009281 | 92.8287 | 86.6534 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 94.4554 | 87.3267 | ovis_aries_rambouillet | ENSOARP00020010811 | 94.4554 | 87.3267 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 93.8614 | 88.3168 | ursus_maritimus | ENSUMAP00000002951 | 83.4507 | 78.5211 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 94.0594 | 87.9208 | ochotona_princeps | ENSOPRP00000015776 | 94.246 | 88.0952 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 95.0495 | 90.297 | delphinapterus_leucas | ENSDLEP00000009945 | 95.0495 | 90.297 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 95.0495 | 90.297 | physeter_catodon | ENSPCTP00005017529 | 91.0816 | 86.5275 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 92.8713 | 86.5347 | mus_caroli | MGP_CAROLIEiJ_P0085506 | 93.4263 | 87.0518 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 92.6733 | 86.7327 | mus_musculus | ENSMUSP00000068380 | 93.2271 | 87.251 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 95.2475 | 91.6832 | dipodomys_ordii | ENSDORP00000008763 | 95.8167 | 92.2311 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 92.8713 | 88.9109 | mustela_putorius_furo | ENSMPUP00000003322 | 82.8622 | 79.3286 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 98.2178 | 97.0297 | aotus_nancymaae | ENSANAP00000038204 | 98.2178 | 97.0297 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 97.6238 | 96.2376 | saimiri_boliviensis_boliviensis | ENSSBOP00000029413 | 94.9904 | 93.6416 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 54.8515 | 53.4653 | vicugna_pacos | ENSVPAP00000002994 | 54.8515 | 53.4653 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 89.505 | 88.9109 | chlorocebus_sabaeus | ENSCSAP00000001611 | 98.4749 | 97.8214 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 94.2574 | 91.8812 | tupaia_belangeri | ENSTBEP00000008412 | 96.945 | 94.501 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 93.4653 | 88.9109 | mesocricetus_auratus | ENSMAUP00000000541 | 94.0239 | 89.4422 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 95.4455 | 90.6931 | phocoena_sinus | ENSPSNP00000022427 | 95.4455 | 90.6931 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 95.2475 | 90.6931 | camelus_dromedarius | ENSCDRP00005002048 | 95.2475 | 90.6931 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 86.5347 | 83.5644 | carlito_syrichta | ENSTSYP00000028281 | 94.5887 | 91.342 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 94.4554 | 90.6931 | panthera_tigris_altaica | ENSPTIP00000015324 | 94.6429 | 90.873 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 90.6931 | 84.7525 | rattus_norvegicus | ENSRNOP00000073704 | 89.4531 | 83.5938 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 90.6931 | 88.1188 | prolemur_simus | ENSPSMP00000009449 | 81.932 | 79.6064 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 92.4753 | 86.7327 | microtus_ochrogaster | ENSMOCP00000012181 | 93.0279 | 87.251 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 86.5347 | 78.6139 | sorex_araneus | ENSSARP00000002078 | 86.7064 | 78.7698 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 92.4753 | 87.3267 | peromyscus_maniculatus_bairdii | ENSPEMP00000024369 | 93.0279 | 87.8486 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 94.6535 | 87.7228 | bos_mutus | ENSBMUP00000013516 | 94.6535 | 87.7228 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 92.8713 | 87.1287 | mus_spicilegus | ENSMSIP00000026321 | 93.4263 | 87.6494 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 92.8713 | 88.5149 | cricetulus_griseus_chok1gshd | ENSCGRP00001012039 | 93.4263 | 89.0438 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 60.9901 | 58.0198 | pteropus_vampyrus | ENSPVAP00000010464 | 61.6 | 58.6 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 72.2772 | 65.9406 | myotis_lucifugus | ENSMLUP00000003514 | 88.8078 | 81.0219 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 80.5941 | 68.3168 | vombatus_ursinus | ENSVURP00010026007 | 81.0757 | 68.7251 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 93.2673 | 87.7228 | rhinolophus_ferrumequinum | ENSRFEP00010026320 | 93.4524 | 87.8968 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 92.2772 | 87.7228 | neovison_vison | ENSNVIP00000021993 | 91.7323 | 87.2047 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 92.6733 | 85.1485 | bos_taurus | ENSBTAP00000065564 | 90.1734 | 82.8516 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 93.0693 | 88.9109 | heterocephalus_glaber_female | ENSHGLP00000053995 | 93.254 | 89.0873 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 86.7327 | 84.7525 | rhinopithecus_bieti | ENSRBIP00000029216 | 95.0108 | 92.8417 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 22.1782 | 20.198 | rhinopithecus_bieti | ENSRBIP00000027300 | 88.189 | 80.315 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 94.6535 | 91.2871 | canis_lupus_dingo | ENSCAFP00020004470 | 87.0674 | 83.9709 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 95.6436 | 91.0891 | sciurus_vulgaris | ENSSVLP00005032739 | 95.8333 | 91.2698 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 80.9901 | 69.3069 | phascolarctos_cinereus | ENSPCIP00000045848 | 81.4741 | 69.7211 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 75.4455 | 69.505 | procavia_capensis | ENSPCAP00000009165 | 82.2894 | 75.8099 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 93.8614 | 89.901 | nannospalax_galili | ENSNGAP00000000629 | 94.4223 | 90.4382 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 94.6535 | 87.7228 | bos_grunniens | ENSBGRP00000038954 | 94.6535 | 87.7228 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 93.4653 | 89.3069 | chinchilla_lanigera | ENSCLAP00000020369 | 93.0966 | 88.9546 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 93.8614 | 86.5347 | cervus_hanglu_yarkandensis | ENSCHYP00000009804 | 93.8614 | 86.5347 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 94.4554 | 90.6931 | catagonus_wagneri | ENSCWAP00000004208 | 94.4554 | 90.6931 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 95.8416 | 91.2871 | ictidomys_tridecemlineatus | ENSSTOP00000022298 | 96.0317 | 91.4683 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 76.2376 | 72.8713 | otolemur_garnettii | ENSOGAP00000004593 | 96.01 | 91.7706 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 98.0198 | 95.6436 | cebus_imitator | ENSCCAP00000016436 | 98.0198 | 95.6436 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 95.8416 | 91.4851 | urocitellus_parryii | ENSUPAP00010017980 | 96.0317 | 91.6667 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 94.0594 | 86.9307 | moschus_moschiferus | ENSMMSP00000013942 | 94.0594 | 86.9307 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 99.0099 | 98.6139 | rhinopithecus_roxellana | ENSRROP00000030919 | 99.0099 | 98.6139 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 92.6733 | 87.3267 | mus_pahari | MGP_PahariEiJ_P0053757 | 93.2271 | 87.8486 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 95.2475 | 92.6733 | propithecus_coquereli | ENSPCOP00000019239 | 92.1456 | 89.6552 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 67.7228 | 53.0693 | callorhinchus_milii | ENSCMIP00000042605 | 66.7969 | 52.3438 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 60.7921 | 48.5149 | latimeria_chalumnae | ENSLACP00000008840 | 72.5768 | 57.9196 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 28.7129 | 23.1683 | lepisosteus_oculatus | ENSLOCP00000008642 | 71.0784 | 57.3529 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 60.7921 | 47.1287 | clupea_harengus | ENSCHAP00000044132 | 48.1191 | 37.3041 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 63.3663 | 47.9208 | danio_rerio | ENSDARP00000107656 | 56.4374 | 42.6808 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 62.7723 | 47.1287 | carassius_auratus | ENSCARP00000068478 | 51.5447 | 38.6992 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 56.6337 | 42.1782 | carassius_auratus | ENSCARP00000025182 | 48.2293 | 35.9191 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 57.2277 | 42.5743 | astyanax_mexicanus | ENSAMXP00000047241 | 51.6995 | 38.4615 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 61.7822 | 46.7327 | ictalurus_punctatus | ENSIPUP00000024879 | 55.2212 | 41.7699 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 62.5743 | 48.3168 | esox_lucius | ENSELUP00000078313 | 45.4023 | 35.0575 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 57.2277 | 43.3663 | electrophorus_electricus | ENSEEEP00000003766 | 58.9796 | 44.6939 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 61.9802 | 47.9208 | oncorhynchus_kisutch | ENSOKIP00005003758 | 48.228 | 37.2881 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 61.9802 | 47.7228 | oncorhynchus_kisutch | ENSOKIP00005060275 | 47.7863 | 36.7939 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 61.9802 | 48.3168 | oncorhynchus_mykiss | ENSOMYP00000135758 | 46.5774 | 36.3095 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 61.3861 | 47.1287 | oncorhynchus_mykiss | ENSOMYP00000044125 | 30.8458 | 23.6816 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 62.1782 | 48.3168 | salmo_salar | ENSSSAP00000153259 | 49.2936 | 38.3046 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 61.7822 | 46.9307 | salmo_salar | ENSSSAP00000143612 | 46.1538 | 35.0592 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 29.703 | 22.1782 | salmo_trutta | ENSSTUP00000006774 | 36.3196 | 27.1186 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 62.3762 | 48.3168 | salmo_trutta | ENSSTUP00000091512 | 48.3871 | 37.4808 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 50.495 | 38.8119 | gadus_morhua | ENSGMOP00000058695 | 50.495 | 38.8119 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 60.9901 | 46.9307 | fundulus_heteroclitus | ENSFHEP00000024517 | 47.3118 | 36.4055 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 57.8218 | 43.7624 | poecilia_reticulata | ENSPREP00000009394 | 41.954 | 31.7529 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 61.3861 | 47.3267 | xiphophorus_maculatus | ENSXMAP00000038288 | 48.5893 | 37.4608 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 60.5941 | 46.7327 | oryzias_latipes | ENSORLP00000017219 | 47.4419 | 36.5891 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 58.0198 | 45.9406 | cyclopterus_lumpus | ENSCLMP00005017054 | 50.1712 | 39.726 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 61.3861 | 47.5248 | oreochromis_niloticus | ENSONIP00000053896 | 47.1125 | 36.4742 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 61.1881 | 47.7228 | haplochromis_burtoni | ENSHBUP00000035378 | 46.9605 | 36.6261 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 61.1881 | 47.7228 | astatotilapia_calliptera | ENSACLP00000024696 | 46.9605 | 36.6261 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 59.802 | 46.1386 | sparus_aurata | ENSSAUP00010034007 | 45.6884 | 35.2496 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 57.4257 | 44.1584 | lates_calcarifer | ENSLCAP00010035753 | 42.09 | 32.3657 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 66.3366 | 52.4752 | xenopus_tropicalis | ENSXETP00000104309 | 65.3021 | 51.6569 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 75.0495 | 61.9802 | chrysemys_picta_bellii | ENSCPBP00000016537 | 74.4597 | 61.4931 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 67.7228 | 55.4455 | sphenodon_punctatus | ENSSPUP00000024303 | 67.9921 | 55.666 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 69.703 | 56.0396 | crocodylus_porosus | ENSCPRP00005004700 | 61.6462 | 49.5622 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 46.3366 | 33.2673 | laticauda_laticaudata | ENSLLTP00000022129 | 56.6586 | 40.678 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 62.7723 | 49.703 | notechis_scutatus | ENSNSUP00000023185 | 66.5966 | 52.7311 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 67.1287 | 53.6634 | pseudonaja_textilis | ENSPTXP00000013721 | 73.6957 | 58.913 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 71.0891 | 58.0198 | anas_platyrhynchos_platyrhynchos | ENSAPLP00000023662 | 62.8722 | 51.3135 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 74.0594 | 59.802 | gallus_gallus | ENSGALP00010030620 | 60.3226 | 48.7097 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 41.9802 | 33.8614 | meleagris_gallopavo | ENSMGAP00000004281 | 69.5082 | 56.0656 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 73.2673 | 57.6238 | serinus_canaria | ENSSCAP00000006223 | 70.8812 | 55.7471 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 73.2673 | 58.2178 | parus_major | ENSPMJP00000002202 | 70.4762 | 56 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 58.6139 | 46.7327 | anolis_carolinensis | ENSACAP00000016205 | 66.9683 | 53.3937 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 58.4158 | 45.7426 | neolamprologus_brichardi | ENSNBRP00000012507 | 48.2815 | 37.8069 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 37.2277 | 29.1089 | naja_naja | ENSNNAP00000028705 | 71.7557 | 56.1069 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 75.0495 | 59.604 | podarcis_muralis | ENSPMRP00000017207 | 74.0234 | 58.7891 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 73.2673 | 58.6139 | geospiza_fortis | ENSGFOP00000003899 | 72.549 | 58.0392 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 73.6634 | 58.6139 | taeniopygia_guttata | ENSTGUP00000032486 | 71.8147 | 57.1429 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 63.3663 | 48.5149 | erpetoichthys_calabaricus | ENSECRP00000021016 | 62.2568 | 47.6654 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 61.7822 | 46.7327 | kryptolebias_marmoratus | ENSKMAP00000022904 | 47.4164 | 35.8663 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 72.6733 | 59.2079 | salvator_merianae | ENSSMRP00000007513 | 70.1721 | 57.1702 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 61.1881 | 46.9307 | oryzias_javanicus | ENSOJAP00000003000 | 48.056 | 36.8585 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 59.2079 | 45.9406 | amphilophus_citrinellus | ENSACIP00000005144 | 55.5762 | 43.1227 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 60.7921 | 46.5347 | dicentrarchus_labrax | ENSDLAP00005081161 | 48.4992 | 37.1248 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 61.1881 | 47.3267 | amphiprion_percula | ENSAPEP00000013668 | 48.8924 | 37.8165 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 60.396 | 46.5347 | oryzias_sinensis | ENSOSIP00000050292 | 46.6361 | 35.9327 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 75.0495 | 61.1881 | pelodiscus_sinensis | ENSPSIP00000008175 | 74.4597 | 60.7073 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 58.6139 | 47.7228 | gopherus_evgoodei | ENSGEVP00005018513 | 71.8447 | 58.4951 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 66.9307 | 54.8515 | chelonoidis_abingdonii | ENSCABP00000028811 | 70.1245 | 57.4689 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 61.3861 | 47.7228 | seriola_dumerili | ENSSDUP00000008262 | 48.1366 | 37.4224 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 60.198 | 46.9307 | cottoperca_gobio | ENSCGOP00000051692 | 48.0253 | 37.4408 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 70.495 | 58.0198 | struthio_camelus_australis | ENSSCUP00000015755 | 74.3215 | 61.1691 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 74.4554 | 60.7921 | anser_brachyrhynchus | ENSABRP00000026389 | 73.7255 | 60.1961 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 48.1188 | 38.8119 | gasterosteus_aculeatus | ENSGACP00000022028 | 60.2978 | 48.6352 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 60.5941 | 46.1386 | cynoglossus_semilaevis | ENSCSEP00000014214 | 47.6636 | 36.2928 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 42.7723 | 34.0594 | poecilia_formosa | ENSPFOP00000016050 | 46.7532 | 37.2294 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 61.7822 | 48.1188 | myripristis_murdjan | ENSMMDP00005052391 | 48.5981 | 37.8505 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 60 | 47.1287 | sander_lucioperca | ENSSLUP00000049718 | 45.9091 | 36.0606 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 38.8119 | 30.297 | strigops_habroptila | ENSSHBP00005005531 | 67.8201 | 52.9412 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 61.3861 | 47.5248 | amphiprion_ocellaris | ENSAOCP00000002458 | 49.0506 | 37.9747 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 75.0495 | 61.7822 | terrapene_carolina_triunguis | ENSTMTP00000003567 | 74.4597 | 61.2967 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 35.2475 | 25.7426 | hucho_hucho | ENSHHUP00000077206 | 37.6321 | 27.4841 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 18.8119 | 16.2376 | hucho_hucho | ENSHHUP00000077213 | 64.6258 | 55.7823 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 47.3267 | 38.4158 | hucho_hucho | ENSHHUP00000002554 | 50.8511 | 41.2766 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 59.802 | 45.9406 | betta_splendens | ENSBSLP00000010502 | 45.8967 | 35.2584 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 62.5743 | 46.7327 | pygocentrus_nattereri | ENSPNAP00000017969 | 53.5593 | 40 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 60.9901 | 46.9307 | anabas_testudineus | ENSATEP00000051866 | 45.7652 | 35.2155 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 55.0495 | 44.1584 | seriola_lalandi_dorsalis | ENSSLDP00000023259 | 59.4017 | 47.6496 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 58.4158 | 44.7525 | labrus_bergylta | ENSLBEP00000015370 | 45.107 | 34.5566 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 60.198 | 46.7327 | pundamilia_nyererei | ENSPNYP00000014715 | 46.2709 | 35.9209 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 61.1881 | 47.3267 | mastacembelus_armatus | ENSMAMP00000061036 | 47.9814 | 37.1118 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 61.3861 | 47.7228 | stegastes_partitus | ENSSPAP00000020149 | 47.1125 | 36.6261 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 44.9505 | 34.4554 | oncorhynchus_tshawytscha | ENSOTSP00005036208 | 53.5377 | 41.0377 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 30.8911 | 21.9802 | oncorhynchus_tshawytscha | ENSOTSP00005036738 | 36.1949 | 25.7541 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 61.3861 | 47.5248 | acanthochromis_polyacanthus | ENSAPOP00000019663 | 47.1125 | 36.4742 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 70.8911 | 56.6337 | aquila_chrysaetos_chrysaetos | ENSACCP00020004834 | 73.8144 | 58.9691 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 53.6634 | 40.7921 | nothobranchius_furzeri | ENSNFUP00015046152 | 40.6907 | 30.9309 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 58.8119 | 45.7426 | cyprinodon_variegatus | ENSCVAP00000013515 | 54.8983 | 42.6987 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 61.5842 | 45.9406 | denticeps_clupeoides | ENSDCDP00000051761 | 48.068 | 35.8578 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 61.1881 | 47.5248 | larimichthys_crocea | ENSLCRP00005063854 | 48.8924 | 37.9747 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 61.7822 | 47.7228 | takifugu_rubripes | ENSTRUP00000048950 | 49.1339 | 37.9528 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 73.4653 | 58.0198 | ficedula_albicollis | ENSFALP00000026149 | 69.7368 | 55.0752 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 60.396 | 46.5347 | hippocampus_comes | ENSHCOP00000000138 | 48.6443 | 37.4801 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 62.7723 | 46.7327 | cyprinus_carpio_carpio | ENSCCRP00000171539 | 50.1582 | 37.3418 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 59.0099 | 42.5743 | cyprinus_carpio_carpio | ENSCCRP00000179567 | 49.584 | 35.7737 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 32.4752 | 26.1386 | coturnix_japonica | ENSCJPP00005014928 | 62.5954 | 50.3817 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 61.3861 | 47.3267 | poecilia_latipinna | ENSPLAP00000008377 | 48.5893 | 37.4608 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 62.5743 | 46.7327 | sinocyclocheilus_grahami | ENSSGRP00000079255 | 54.1096 | 40.411 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 57.2277 | 42.1782 | sinocyclocheilus_grahami | ENSSGRP00000037583 | 51.6995 | 38.1038 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 61.3861 | 47.7228 | maylandia_zebra | ENSMZEP00005008903 | 47.1125 | 36.6261 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 47.7228 | 36.6337 | tetraodon_nigroviridis | ENSTNIP00000016611 | 60.5528 | 46.4824 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 62.3762 | 46.9307 | paramormyrops_kingsleyae | ENSPKIP00000010808 | 54.5927 | 41.0745 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 61.1881 | 46.7327 | scleropages_formosus | ENSSFOP00015001668 | 55.7762 | 42.5993 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 60.198 | 47.5248 | leptobrachium_leishanense | ENSLLEP00000024125 | 67.2566 | 53.0973 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 60.5941 | 46.7327 | scophthalmus_maximus | ENSSMAP00000014555 | 49.0385 | 37.8205 |
| ENSG00000140718 | homo_sapiens | ENSP00000418823 | 61.1881 | 46.9307 | oryzias_melastigma | ENSOMEP00000014338 | 48.1308 | 36.9159 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0001659 | involved_in temperature homeostasis | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0005634 | located_in nucleus | 2 | IDA,ISS | Homo_sapiens(9606) | Component |
| GO:0005654 | located_in nucleoplasm | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0005737 | located_in cytoplasm | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005829 | located_in cytosol | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005886 | located_in plasma membrane | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0006307 | involved_in DNA dealkylation involved in DNA repair | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0008198 | enables ferrous iron binding | 2 | IBA,IDA | Homo_sapiens(9606) | Function |
| GO:0010883 | involved_in regulation of lipid storage | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0016607 | located_in nuclear speck | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0016740 | enables transferase activity | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0035515 | enables oxidative RNA demethylase activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0035516 | enables oxidative DNA demethylase activity | 2 | IBA,IDA | Homo_sapiens(9606) | Function |
| GO:0035552 | involved_in oxidative single-stranded DNA demethylation | 2 | IBA,IDA | Homo_sapiens(9606) | Process |
| GO:0035553 | involved_in oxidative single-stranded RNA demethylation | 3 | IBA,IDA,IMP | Homo_sapiens(9606) | Process |
| GO:0040014 | involved_in regulation of multicellular organism growth | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0042245 | involved_in RNA repair | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0043231 | located_in intracellular membrane-bounded organelle | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0043734 | enables DNA-N1-methyladenine dioxygenase activity | 2 | IBA,IDA | Homo_sapiens(9606) | Function |
| GO:0044065 | involved_in regulation of respiratory system process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0060612 | involved_in adipose tissue development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0061157 | involved_in mRNA destabilization | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0070350 | involved_in regulation of white fat cell proliferation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0070989 | involved_in oxidative demethylation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0080111 | involved_in DNA demethylation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0090335 | involved_in regulation of brown fat cell differentiation | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:1990931 | enables mRNA N6-methyladenosine dioxygenase activity | 3 | IBA,IDA,IMP | Homo_sapiens(9606) | Function |
| GO:1990984 | enables tRNA demethylase activity | 1 | IDA | Homo_sapiens(9606) | Function |