Welcome to
RBP World!
Description
| Ensembl ID | ENSG00000136807 | Gene ID | 1025 | Accession | 1780 |
| Symbol | CDK9 | Alias | TAK;C-2k;CTK1;CDC2L4;PITALRE | Full Name | cyclin dependent kinase 9 |
| Status | Confidence | Length | 5114 bases | Strand | Plus strand |
| Position | 9 : 127785679 - 127790792 | RNA binding domain | N.A. | ||
| Summary | The protein encoded by this gene is a member of the cyclin-dependent protein kinase (CDK) family. CDK family members are highly similar to the gene products of S. cerevisiae cdc28, and S. pombe cdc2, and known as important cell cycle regulators. This kinase was found to be a component of the multiprotein complex TAK/P-TEFb, which is an elongation factor for RNA polymerase II-directed transcription and functions by phosphorylating the C-terminal domain of the largest subunit of RNA polymerase II. This protein forms a complex with and is regulated by its regulatory subunit cyclin T or cyclin K. HIV-1 Tat protein was found to interact with this protein and cyclin T, which suggested a possible involvement of this protein in AIDS. [provided by RefSeq, Jul 2008] | ||||
RNA binding domains (RBDs)
| Protein | Domain | Pfam ID | E-value | Domain number |
|---|
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|---|---|---|---|
| 33717648 | Lnc(ing)RNAs to the 'shock and kill' strategy for HIV-1 cure. | Saikat Boliar | 2021-03-05 | Molecular therapy. Nucleic acids |
| 24694320 | A novel, non-radioactive eukaryotic in vitro transcription assay for sensitive quantification of RNA polymerase II activity. | Cristina Voss | 2014-04-03 | BMC molecular biology |
| 31020497 | Tat expression led to increased histone 3 tri-methylation at lysine 27 and contributed to HIV latency in astrocytes through regulation of MeCP2 and Ezh2 expression. | Ying Liu | 2019-08-01 | Journal of neurovirology |
| 21205204 | Phosphorylation mechanism and structure of serine-arginine protein kinases. | Gourisankar Ghosh | 2011-02-01 | The FEBS journal |
| 22672903 | A role for CDK9 in UV damage response. | E Magdalena Morawska | 2012-06-15 | Cell cycle (Georgetown, Tex.) |
| 31953354 | IL-1 Transcriptional Responses to Lipopolysaccharides Are Regulated by a Complex of RNA Binding Proteins. | Lihua Shi | 2020-03-01 | Journal of immunology (Baltimore, Md. : 1950) |
| 35693926 | Inhibition of Cyclin-Dependent Kinase 9 Downregulates Cytokine Production Without Detrimentally Affecting Human Monocyte-Derived Macrophage Viability. | J Brian McHugh | 2022-01-01 | Frontiers in cell and developmental biology |
| 32778962 | Repurposing of Kinase Inhibitors for Treatment of COVID-19. | Ellen Weisberg | 2020-08-10 | Pharmaceutical research |
| 27171036 | Cyclin Dependent Kinase 9 Inhibitors for Cancer Therapy. | A Yogesh Sonawane | 2016-10-13 | Journal of medicinal chemistry |
| 22140483 | A flavonoid, luteolin, cripples HIV-1 by abrogation of tat function. | Rajeev Mehla | 2011-01-01 | PloS one |
| 35856391 | hnRNP R negatively regulates transcription by modulating the association of P-TEFb with 7SK and BRD4. | Changhe Ji | 2022-09-05 | EMBO reports |
| 30080437 | JQ1 affects BRD2-dependent and independent transcription regulation without disrupting H4-hyperacetylated chromatin states. | Lusy Handoko | 2018-01-01 | Epigenetics |
| 30613326 | Privileged Structures and Polypharmacology within and between Protein Families. | Joshua Meyers | 2018-12-13 | ACS medicinal chemistry letters |
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| CDK9-202 | ENST00000421939 | 702 | 194aa | ENSP00000395872 |
| CDK9-201 | ENST00000373264 | 2483 | 372aa | ENSP00000362361 |
| CDK9-203 | ENST00000480353 | 706 | No protein | - |
| CDK9-204 | ENST00000491521 | 510 | No protein | - |
| CDK9-205 | ENST00000498339 | 458 | No protein | - |
Phenotypes
| ensgID | Trait | pValue | Pubmed ID |
|---|
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
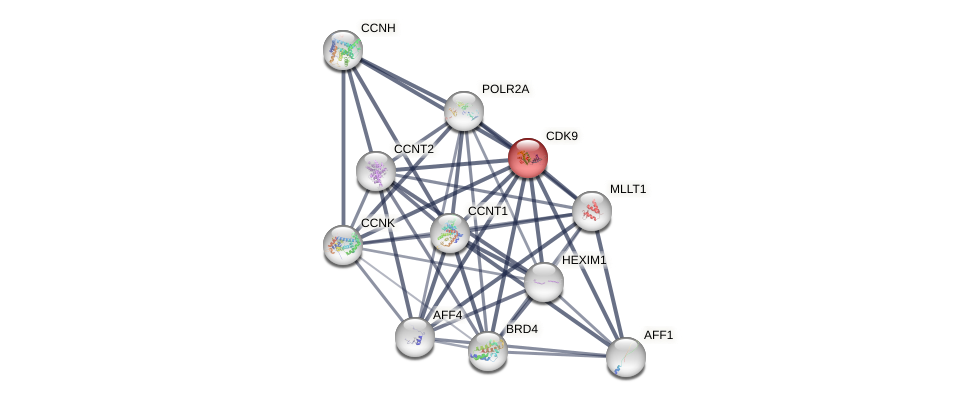
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000136807 | homo_sapiens | ENSP00000337194 | 14.1013 | 8.54022 | homo_sapiens | ENSP00000362361 | 38.172 | 23.1183 |
| ENSG00000136807 | homo_sapiens | ENSP00000181839 | 15.6746 | 11.1111 | homo_sapiens | ENSP00000362361 | 63.7097 | 45.1613 |
| ENSG00000136807 | homo_sapiens | ENSP00000498608 | 39.3103 | 27.3563 | homo_sapiens | ENSP00000362361 | 45.9677 | 31.9892 |
| ENSG00000136807 | homo_sapiens | ENSP00000485244 | 17.6042 | 11.4583 | homo_sapiens | ENSP00000362361 | 45.4301 | 29.5699 |
| ENSG00000136807 | homo_sapiens | ENSP00000261211 | 35.9465 | 22.3709 | homo_sapiens | ENSP00000362361 | 50.5376 | 31.4516 |
| ENSG00000136807 | homo_sapiens | ENSP00000370938 | 38.5776 | 25.431 | homo_sapiens | ENSP00000362361 | 48.1183 | 31.7204 |
| ENSG00000136807 | homo_sapiens | ENSP00000266970 | 56.7114 | 39.2617 | homo_sapiens | ENSP00000362361 | 45.4301 | 31.4516 |
| ENSG00000136807 | homo_sapiens | ENSP00000322343 | 47.9769 | 34.3931 | homo_sapiens | ENSP00000362361 | 44.6237 | 31.9892 |
| ENSG00000136807 | homo_sapiens | ENSP00000349762 | 37.9032 | 26.0081 | homo_sapiens | ENSP00000362361 | 50.5376 | 34.6774 |
| ENSG00000136807 | homo_sapiens | ENSP00000338673 | 48.8889 | 33.3333 | homo_sapiens | ENSP00000362361 | 47.3118 | 32.2581 |
| ENSG00000136807 | homo_sapiens | ENSP00000463048 | 24.0409 | 15.4731 | homo_sapiens | ENSP00000362361 | 50.5376 | 32.5269 |
| ENSG00000136807 | homo_sapiens | ENSP00000384442 | 24.2308 | 15.5128 | homo_sapiens | ENSP00000362361 | 50.8064 | 32.5269 |
| ENSG00000136807 | homo_sapiens | ENSP00000257904 | 52.8053 | 34.9835 | homo_sapiens | ENSP00000362361 | 43.0108 | 28.4946 |
| ENSG00000136807 | homo_sapiens | ENSP00000306340 | 31.2281 | 18.4211 | homo_sapiens | ENSP00000362361 | 47.8495 | 28.2258 |
| ENSG00000136807 | homo_sapiens | ENSP00000256443 | 50.578 | 35.5491 | homo_sapiens | ENSP00000362361 | 47.043 | 33.0645 |
| ENSG00000136807 | homo_sapiens | ENSP00000398880 | 16.0403 | 11.2752 | homo_sapiens | ENSP00000362361 | 64.2473 | 45.1613 |
| ENSG00000136807 | homo_sapiens | ENSP00000265334 | 29.5608 | 17.9054 | homo_sapiens | ENSP00000362361 | 47.043 | 28.4946 |
| ENSG00000136807 | homo_sapiens | ENSP00000397087 | 52.454 | 34.0491 | homo_sapiens | ENSP00000362361 | 45.9677 | 29.8387 |
| ENSG00000136807 | homo_sapiens | ENSP00000514486 | 48.8304 | 30.117 | homo_sapiens | ENSP00000362361 | 44.8925 | 27.6882 |
| ENSG00000136807 | homo_sapiens | ENSP00000419782 | 57.1918 | 40.0685 | homo_sapiens | ENSP00000362361 | 44.8925 | 31.4516 |
| ENSG00000136807 | homo_sapiens | ENSP00000399082 | 38.1857 | 26.1603 | homo_sapiens | ENSP00000362361 | 48.6559 | 33.3333 |
| ENSG00000136807 | homo_sapiens | ENSP00000369390 | 37.7399 | 25.1599 | homo_sapiens | ENSP00000362361 | 47.5806 | 31.7204 |
| ENSG00000136807 | homo_sapiens | ENSP00000400088 | 55.082 | 38.3607 | homo_sapiens | ENSP00000362361 | 45.1613 | 31.4516 |
| ENSG00000136807 | homo_sapiens | ENSP00000378699 | 58.5859 | 39.7306 | homo_sapiens | ENSP00000362361 | 46.7742 | 31.7204 |
| ENSG00000136807 | homo_sapiens | ENSP00000379176 | 48.7395 | 30.5322 | homo_sapiens | ENSP00000362361 | 46.7742 | 29.3011 |
| ENSG00000136807 | homo_sapiens | ENSP00000357907 | 35.8566 | 23.9044 | homo_sapiens | ENSP00000362361 | 48.3871 | 32.2581 |
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 96.7742 | 95.9677 | nomascus_leucogenys | ENSNLEP00000021235 | 96.7742 | 95.9677 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 100 | 100 | pan_paniscus | ENSPPAP00000024837 | 100 | 100 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.7312 | 99.7312 | pongo_abelii | ENSPPYP00000021999 | 75.7143 | 75.7143 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 100 | 100 | pan_troglodytes | ENSPTRP00000036573 | 100 | 100 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 100 | 100 | gorilla_gorilla | ENSGGOP00000010772 | 100 | 100 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 79.0323 | 72.3118 | ornithorhynchus_anatinus | ENSOANP00000040556 | 75 | 68.6225 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 95.6989 | 92.7419 | sarcophilus_harrisii | ENSSHAP00000037663 | 70.495 | 68.3168 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 73.3871 | 70.6989 | notamacropus_eugenii | ENSMEUP00000011890 | 59.4771 | 57.2985 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 98.9247 | 97.5806 | dasypus_novemcinctus | ENSDNOP00000032730 | 75.5647 | 74.538 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 58.6021 | 58.0645 | erinaceus_europaeus | ENSEEUP00000012009 | 52.0286 | 51.5513 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 90.3226 | 87.9032 | echinops_telfairi | ENSETEP00000006847 | 68.9938 | 67.1458 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.7312 | 98.9247 | callithrix_jacchus | ENSCJAP00000036928 | 76.1807 | 75.5647 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 100 | 100 | cercocebus_atys | ENSCATP00000037570 | 100 | 100 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 100 | 100 | macaca_fascicularis | ENSMFAP00000035326 | 100 | 100 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 100 | 100 | macaca_mulatta | ENSMMUP00000073882 | 100 | 100 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 100 | 100 | macaca_nemestrina | ENSMNEP00000031396 | 100 | 100 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 100 | 100 | papio_anubis | ENSPANP00000019372 | 76.2295 | 76.2295 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 100 | 100 | mandrillus_leucophaeus | ENSMLEP00000003030 | 100 | 100 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 98.1183 | 97.043 | tursiops_truncatus | ENSTTRP00000015605 | 74.9487 | 74.1273 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.4624 | 98.1183 | vulpes_vulpes | ENSVVUP00000018023 | 99.4624 | 98.1183 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.7312 | 98.6559 | mus_spretus | MGP_SPRETEiJ_P0053226 | 99.7312 | 98.6559 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.4624 | 98.3871 | equus_asinus | ENSEASP00005049589 | 70.2087 | 69.4497 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.7312 | 98.6559 | capra_hircus | ENSCHIP00000029772 | 99.7312 | 98.6559 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.4624 | 98.3871 | loxodonta_africana | ENSLAFP00000014128 | 99.4624 | 98.3871 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.4624 | 97.8495 | panthera_leo | ENSPLOP00000027001 | 75.3564 | 74.1344 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.1936 | 97.8495 | ailuropoda_melanoleuca | ENSAMEP00000023903 | 66.129 | 65.233 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.7312 | 98.6559 | bos_indicus_hybrid | ENSBIXP00005011546 | 75.1012 | 74.2915 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.4624 | 98.6559 | equus_caballus | ENSECAP00000036625 | 74.8988 | 74.2915 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.4624 | 98.1183 | canis_lupus_familiaris | ENSCAFP00845007462 | 75.6646 | 74.6421 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.4624 | 97.8495 | panthera_pardus | ENSPPRP00000020097 | 99.4624 | 97.8495 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.4624 | 98.3871 | marmota_marmota_marmota | ENSMMMP00000019475 | 99.4624 | 98.3871 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.4624 | 98.6559 | balaenoptera_musculus | ENSBMSP00010005840 | 75.8197 | 75.2049 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.7312 | 98.3871 | cavia_porcellus | ENSCPOP00000009305 | 99.7312 | 98.3871 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 79.0323 | 73.1183 | cavia_porcellus | ENSCPOP00000008151 | 87.2404 | 80.7122 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.4624 | 97.8495 | felis_catus | ENSFCAP00000028744 | 75.2033 | 73.9837 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.1936 | 97.8495 | ursus_americanus | ENSUAMP00000029754 | 99.1936 | 97.8495 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 95.6989 | 92.7419 | monodelphis_domestica | ENSMODP00000040851 | 70.0787 | 67.9134 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.7312 | 98.6559 | bison_bison_bison | ENSBBBP00000025566 | 99.7312 | 98.6559 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.1936 | 98.3871 | monodon_monoceros | ENSMMNP00015028874 | 99.1936 | 98.3871 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.4624 | 98.6559 | sus_scrofa | ENSSSCP00000006022 | 99.4624 | 98.6559 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.4624 | 98.3871 | microcebus_murinus | ENSMICP00000043693 | 99.4624 | 98.3871 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.4624 | 98.3871 | octodon_degus | ENSODEP00000005231 | 99.4624 | 98.3871 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.7312 | 98.3871 | jaculus_jaculus | ENSJJAP00000014867 | 99.7312 | 98.3871 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.7312 | 98.6559 | ovis_aries_rambouillet | ENSOARP00020020388 | 99.7312 | 98.6559 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.0107 | 90.8602 | ursus_maritimus | ENSUMAP00000000625 | 93.0107 | 90.8602 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.1936 | 98.3871 | delphinapterus_leucas | ENSDLEP00000019389 | 75.6148 | 75 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.4624 | 98.3871 | physeter_catodon | ENSPCTP00005012225 | 99.4624 | 98.3871 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.7312 | 98.6559 | mus_caroli | MGP_CAROLIEiJ_P0051186 | 99.7312 | 98.6559 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.7312 | 98.6559 | mus_musculus | ENSMUSP00000009699 | 99.7312 | 98.6559 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.4624 | 97.8495 | dipodomys_ordii | ENSDORP00000012530 | 99.4624 | 97.8495 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.4624 | 97.8495 | mustela_putorius_furo | ENSMPUP00000013578 | 99.4624 | 97.8495 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.2796 | 91.9355 | aotus_nancymaae | ENSANAP00000040868 | 96.3889 | 95 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 88.9785 | 85.7527 | aotus_nancymaae | ENSANAP00000026018 | 94.0341 | 90.625 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.7312 | 98.9247 | aotus_nancymaae | ENSANAP00000042080 | 99.7312 | 98.9247 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 85.2151 | 80.914 | saimiri_boliviensis_boliviensis | ENSSBOP00000027122 | 86.612 | 82.2404 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.7312 | 98.9247 | saimiri_boliviensis_boliviensis | ENSSBOP00000013289 | 99.7312 | 98.9247 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 62.0968 | 61.2903 | vicugna_pacos | ENSVPAP00000004470 | 67.9412 | 67.0588 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 100 | 100 | chlorocebus_sabaeus | ENSCSAP00000009958 | 76.2295 | 76.2295 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.7312 | 98.1183 | mesocricetus_auratus | ENSMAUP00000023295 | 99.7312 | 98.1183 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.4624 | 98.6559 | phocoena_sinus | ENSPSNP00000014199 | 75.8197 | 75.2049 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.4624 | 98.6559 | camelus_dromedarius | ENSCDRP00005018647 | 99.4624 | 98.6559 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 91.129 | 90.5914 | carlito_syrichta | ENSTSYP00000017054 | 99.4135 | 98.827 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 91.129 | 89.7849 | panthera_tigris_altaica | ENSPTIP00000001850 | 91.129 | 89.7849 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.7312 | 98.6559 | rattus_norvegicus | ENSRNOP00000037778 | 75.5601 | 74.7454 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.7312 | 98.9247 | prolemur_simus | ENSPSMP00000011597 | 99.7312 | 98.9247 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.7312 | 98.6559 | microtus_ochrogaster | ENSMOCP00000026218 | 99.7312 | 98.6559 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.7312 | 98.6559 | peromyscus_maniculatus_bairdii | ENSPEMP00000008964 | 99.7312 | 98.6559 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.7312 | 98.6559 | bos_mutus | ENSBMUP00000025255 | 99.7312 | 98.6559 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.7312 | 98.6559 | mus_spicilegus | ENSMSIP00000011770 | 76.1807 | 75.3593 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.7312 | 98.6559 | cricetulus_griseus_chok1gshd | ENSCGRP00001013357 | 75.8691 | 75.0511 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.1936 | 98.1183 | pteropus_vampyrus | ENSPVAP00000000911 | 76.0825 | 75.2577 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 97.3118 | 95.9677 | myotis_lucifugus | ENSMLUP00000012271 | 97.0509 | 95.7105 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.5484 | 89.7849 | myotis_lucifugus | ENSMLUP00000013177 | 93.5484 | 89.7849 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 91.129 | 86.828 | myotis_lucifugus | ENSMLUP00000019184 | 91.129 | 86.828 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 94.8925 | 92.2043 | myotis_lucifugus | ENSMLUP00000019093 | 94.8925 | 92.2043 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 91.6667 | 89.2473 | myotis_lucifugus | ENSMLUP00000021932 | 91.6667 | 89.2473 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 95.6989 | 92.7419 | vombatus_ursinus | ENSVURP00010021035 | 95.4424 | 92.4933 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.1936 | 98.1183 | rhinolophus_ferrumequinum | ENSRFEP00010023029 | 75.6148 | 74.7951 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.4624 | 97.8495 | neovison_vison | ENSNVIP00000013180 | 99.4624 | 97.8495 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.7312 | 98.6559 | bos_taurus | ENSBTAP00000006162 | 99.7312 | 98.6559 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.4624 | 97.8495 | heterocephalus_glaber_female | ENSHGLP00000017562 | 75.9754 | 74.7433 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 100 | 100 | rhinopithecus_bieti | ENSRBIP00000030488 | 100 | 100 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.4624 | 98.1183 | canis_lupus_dingo | ENSCAFP00020019882 | 75.6646 | 74.6421 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.4624 | 98.1183 | sciurus_vulgaris | ENSSVLP00005017987 | 76.1317 | 75.1029 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 65.3226 | 64.2473 | procavia_capensis | ENSPCAP00000008289 | 69.8276 | 68.6782 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.7312 | 98.9247 | nannospalax_galili | ENSNGAP00000017885 | 99.7312 | 98.9247 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.7312 | 98.6559 | bos_grunniens | ENSBGRP00000010546 | 75.1012 | 74.2915 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 97.8495 | 97.043 | chinchilla_lanigera | ENSCLAP00000012127 | 97.8495 | 97.043 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.7312 | 98.6559 | cervus_hanglu_yarkandensis | ENSCHYP00000027381 | 99.7312 | 98.6559 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.7312 | 98.9247 | catagonus_wagneri | ENSCWAP00000014978 | 76.0246 | 75.4098 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.4624 | 98.3871 | ictidomys_tridecemlineatus | ENSSTOP00000007412 | 99.4624 | 98.3871 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.4624 | 98.6559 | otolemur_garnettii | ENSOGAP00000007253 | 99.4624 | 98.6559 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.7312 | 98.9247 | cebus_imitator | ENSCCAP00000032481 | 99.7312 | 98.9247 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 82.7957 | 80.1075 | cebus_imitator | ENSCCAP00000013882 | 93.0514 | 90.0302 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.4624 | 98.3871 | urocitellus_parryii | ENSUPAP00010018088 | 76.1317 | 75.3086 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.7312 | 98.6559 | moschus_moschiferus | ENSMMSP00000027753 | 99.7312 | 98.6559 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 100 | 100 | rhinopithecus_roxellana | ENSRROP00000034594 | 100 | 100 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.7312 | 98.6559 | mus_pahari | MGP_PahariEiJ_P0062197 | 99.7312 | 98.6559 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 99.4624 | 98.3871 | propithecus_coquereli | ENSPCOP00000023785 | 99.4624 | 98.3871 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 70.6989 | 53.7634 | caenorhabditis_elegans | H25P06.2b.1 | 55.0209 | 41.841 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 83.0645 | 71.2366 | drosophila_melanogaster | FBpp0071848 | 76.4851 | 65.5941 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 82.5269 | 73.3871 | ciona_intestinalis | ENSCINP00000018002 | 81.6489 | 72.6064 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 39.5161 | 36.5591 | petromyzon_marinus | ENSPMAP00000006866 | 86.9822 | 80.4734 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 89.5161 | 83.3333 | eptatretus_burgeri | ENSEBUP00000017260 | 88.5638 | 82.4468 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 81.4516 | 76.6129 | callorhinchus_milii | ENSCMIP00000034259 | 88.0814 | 82.8488 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 94.6237 | 90.0538 | latimeria_chalumnae | ENSLACP00000019397 | 94.6237 | 90.0538 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 70.9677 | 68.8172 | lepisosteus_oculatus | ENSLOCP00000006398 | 92.9577 | 90.1408 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.2796 | 88.4409 | clupea_harengus | ENSCHAP00000013195 | 92.5333 | 87.7333 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.8172 | 89.5161 | danio_rerio | ENSDARP00000065858 | 88.8041 | 84.7328 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.5484 | 88.4409 | carassius_auratus | ENSCARP00000041450 | 88.5496 | 83.715 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 92.4731 | 87.0968 | carassius_auratus | ENSCARP00000068572 | 88.8889 | 83.7209 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.5484 | 88.4409 | carassius_auratus | ENSCARP00000085800 | 88.5496 | 83.715 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 90.5914 | 84.4086 | carassius_auratus | ENSCARP00000069122 | 87.3057 | 81.3472 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.2796 | 88.4409 | astyanax_mexicanus | ENSAMXP00000013753 | 75.93 | 71.9912 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 92.7419 | 87.0968 | ictalurus_punctatus | ENSIPUP00000031698 | 87.5635 | 82.2335 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.0107 | 88.172 | ictalurus_punctatus | ENSIPUP00000031655 | 87.8173 | 83.2487 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 91.6667 | 86.0215 | esox_lucius | ENSELUP00000039429 | 87.6607 | 82.2622 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.2796 | 88.9785 | electrophorus_electricus | ENSEEEP00000025235 | 92.5333 | 88.2667 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.2796 | 88.7097 | oncorhynchus_kisutch | ENSOKIP00005088932 | 92.7807 | 88.2353 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.2796 | 88.7097 | oncorhynchus_mykiss | ENSOMYP00000091517 | 92.7807 | 88.2353 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.2796 | 88.9785 | salmo_salar | ENSSSAP00000090031 | 88.2952 | 84.2239 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.2796 | 88.7097 | salmo_trutta | ENSSTUP00000039805 | 92.7807 | 88.2353 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 92.7419 | 88.172 | gadus_morhua | ENSGMOP00000063994 | 80.6075 | 76.6355 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.5484 | 88.4409 | fundulus_heteroclitus | ENSFHEP00000023288 | 83.0549 | 78.5203 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 82.5269 | 75.8064 | poecilia_reticulata | ENSPREP00000011203 | 55.6159 | 51.087 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.0107 | 88.172 | xiphophorus_maculatus | ENSXMAP00000003851 | 88.0407 | 83.4606 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.0107 | 88.9785 | oryzias_latipes | ENSORLP00000022884 | 88.0407 | 84.2239 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.5484 | 88.7097 | cyclopterus_lumpus | ENSCLMP00005028205 | 88.5496 | 83.9695 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.5484 | 88.9785 | oreochromis_niloticus | ENSONIP00000015861 | 88.5496 | 84.2239 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.5484 | 88.9785 | haplochromis_burtoni | ENSHBUP00000029062 | 88.5496 | 84.2239 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.5484 | 88.9785 | astatotilapia_calliptera | ENSACLP00000007818 | 88.5496 | 84.2239 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.0107 | 88.4409 | sparus_aurata | ENSSAUP00010018557 | 85.6436 | 81.4356 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.5484 | 88.7097 | lates_calcarifer | ENSLCAP00010025072 | 93.0481 | 88.2353 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 94.3548 | 89.7849 | xenopus_tropicalis | ENSXETP00000031527 | 93.3511 | 88.8298 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 94.8925 | 91.9355 | chrysemys_picta_bellii | ENSCPBP00000036289 | 94.8925 | 91.9355 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 90.3226 | 86.828 | sphenodon_punctatus | ENSSPUP00000023742 | 89.1247 | 85.6764 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 94.086 | 91.3979 | crocodylus_porosus | ENSCPRP00005000326 | 86.2069 | 83.7438 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 94.086 | 90.3226 | laticauda_laticaudata | ENSLLTP00000004968 | 94.086 | 90.3226 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 88.9785 | 84.6774 | notechis_scutatus | ENSNSUP00000000742 | 89.9457 | 85.5978 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 94.6237 | 91.129 | pseudonaja_textilis | ENSPTXP00000010979 | 86.9136 | 83.7037 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 87.6344 | 84.9462 | anas_platyrhynchos_platyrhynchos | ENSAPLP00000028577 | 80.8933 | 78.4119 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 94.3548 | 91.9355 | gallus_gallus | ENSGALP00010041820 | 80.8756 | 78.8018 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 88.7097 | 84.9462 | meleagris_gallopavo | ENSMGAP00000003475 | 76.5661 | 73.3179 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 94.6237 | 92.2043 | serinus_canaria | ENSSCAP00000008442 | 85.0242 | 82.8502 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 94.6237 | 92.2043 | parus_major | ENSPMJP00000012415 | 94.6237 | 92.2043 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.5484 | 88.9785 | neolamprologus_brichardi | ENSNBRP00000005387 | 93.5484 | 88.9785 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 86.828 | 82.5269 | naja_naja | ENSNNAP00000029686 | 88.4931 | 84.1096 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 94.3548 | 90.8602 | podarcis_muralis | ENSPMRP00000025434 | 86.6667 | 83.4568 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 88.4409 | 86.0215 | geospiza_fortis | ENSGFOP00000006038 | 89.6458 | 87.1935 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 94.6237 | 92.2043 | taeniopygia_guttata | ENSTGUP00000003698 | 90.0256 | 87.7238 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 87.3656 | 83.6021 | erpetoichthys_calabaricus | ENSECRP00000024591 | 89.2857 | 85.4396 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.0107 | 88.9785 | kryptolebias_marmoratus | ENSKMAP00000007518 | 88.0407 | 84.2239 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 94.3548 | 91.129 | salvator_merianae | ENSSMRP00000014314 | 94.3548 | 91.129 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.5484 | 89.2473 | oryzias_javanicus | ENSOJAP00000031092 | 88.5496 | 84.4784 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.5484 | 88.7097 | amphilophus_citrinellus | ENSACIP00000015697 | 93.0481 | 88.2353 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.2796 | 88.7097 | dicentrarchus_labrax | ENSDLAP00005027391 | 88.2952 | 83.9695 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.5484 | 88.7097 | amphiprion_percula | ENSAPEP00000015884 | 88.5496 | 83.9695 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 59.6774 | 56.4516 | oryzias_sinensis | ENSOSIP00000003244 | 88.8 | 84 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 88.9785 | 85.2151 | pelodiscus_sinensis | ENSPSIP00000013666 | 89.7019 | 85.9079 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 94.8925 | 91.9355 | gopherus_evgoodei | ENSGEVP00005026932 | 94.8925 | 91.9355 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 94.8925 | 91.9355 | chelonoidis_abingdonii | ENSCABP00000002309 | 94.8925 | 91.9355 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.5484 | 88.9785 | seriola_dumerili | ENSSDUP00000025736 | 88.5496 | 84.2239 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.5484 | 88.9785 | cottoperca_gobio | ENSCGOP00000030741 | 88.5496 | 84.2239 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 87.0968 | 83.871 | struthio_camelus_australis | ENSSCUP00000023396 | 91.0112 | 87.6404 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 94.086 | 91.3979 | anser_brachyrhynchus | ENSABRP00000000637 | 94.086 | 91.3979 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.5484 | 88.9785 | gasterosteus_aculeatus | ENSGACP00000024480 | 93.5484 | 88.9785 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 92.7419 | 88.7097 | cynoglossus_semilaevis | ENSCSEP00000015411 | 87.7863 | 83.9695 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.2796 | 88.4409 | poecilia_formosa | ENSPFOP00000005331 | 88.2952 | 83.715 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 56.9892 | 49.1936 | myripristis_murdjan | ENSMMDP00005003689 | 69.7368 | 60.1974 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.5484 | 88.9785 | myripristis_murdjan | ENSMMDP00005006358 | 93.0481 | 88.5027 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.2796 | 88.4409 | sander_lucioperca | ENSSLUP00000046317 | 87.1859 | 82.6633 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 94.3548 | 92.2043 | strigops_habroptila | ENSSHBP00005019116 | 94.3548 | 92.2043 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.5484 | 88.7097 | amphiprion_ocellaris | ENSAOCP00000024696 | 88.5496 | 83.9695 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 94.8925 | 91.9355 | terrapene_carolina_triunguis | ENSTMTP00000029974 | 94.8925 | 91.9355 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.2796 | 88.9785 | hucho_hucho | ENSHHUP00000080583 | 92.7807 | 88.5027 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.5484 | 88.7097 | betta_splendens | ENSBSLP00000033679 | 88.5496 | 83.9695 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.2796 | 88.9785 | pygocentrus_nattereri | ENSPNAP00000022944 | 88.5204 | 84.4388 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.2796 | 87.9032 | anabas_testudineus | ENSATEP00000010877 | 88.2952 | 83.2061 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 82.7957 | 73.6559 | ciona_savignyi | ENSCSAVP00000011226 | 82.3529 | 73.262 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 57.5269 | 37.6344 | saccharomyces_cerevisiae | YPR161C | 32.5723 | 21.309 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.5484 | 88.7097 | seriola_lalandi_dorsalis | ENSSLDP00000022391 | 93.0481 | 88.2353 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.0107 | 88.4409 | labrus_bergylta | ENSLBEP00000028021 | 93.0107 | 88.4409 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.5484 | 88.9785 | pundamilia_nyererei | ENSPNYP00000004383 | 88.5496 | 84.2239 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.5484 | 88.9785 | mastacembelus_armatus | ENSMAMP00000062700 | 79.6339 | 75.7437 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.2796 | 88.7097 | stegastes_partitus | ENSSPAP00000015409 | 88.2952 | 83.9695 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.5484 | 88.9785 | acanthochromis_polyacanthus | ENSAPOP00000010283 | 93.5484 | 88.9785 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 94.086 | 91.3979 | aquila_chrysaetos_chrysaetos | ENSACCP00020024825 | 94.086 | 91.3979 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 92.4731 | 87.9032 | nothobranchius_furzeri | ENSNFUP00015002748 | 87.5318 | 83.2061 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.2796 | 88.4409 | cyprinodon_variegatus | ENSCVAP00000003473 | 93.2796 | 88.4409 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 92.4731 | 87.6344 | denticeps_clupeoides | ENSDCDP00000047864 | 87.5318 | 82.9517 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.2796 | 88.4409 | larimichthys_crocea | ENSLCRP00005009540 | 93.0295 | 88.2038 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.0107 | 88.4409 | takifugu_rubripes | ENSTRUP00000022568 | 88.2653 | 83.9286 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 94.6237 | 92.2043 | ficedula_albicollis | ENSFALP00000001395 | 76.3558 | 74.4035 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.0107 | 88.9785 | hippocampus_comes | ENSHCOP00000001526 | 85.8561 | 82.134 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.5484 | 88.4409 | cyprinus_carpio_carpio | ENSCCRP00000112367 | 88.3249 | 83.5025 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.5484 | 88.4409 | cyprinus_carpio_carpio | ENSCCRP00000133742 | 88.5496 | 83.715 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 94.3548 | 91.9355 | coturnix_japonica | ENSCJPP00005025394 | 94.3548 | 91.9355 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.2796 | 88.4409 | poecilia_latipinna | ENSPLAP00000016154 | 88.2952 | 83.715 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.5484 | 88.172 | sinocyclocheilus_grahami | ENSSGRP00000064053 | 88.5496 | 83.4606 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.5484 | 88.9785 | maylandia_zebra | ENSMZEP00005001044 | 88.5496 | 84.2239 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 78.7634 | 75.5376 | tetraodon_nigroviridis | ENSTNIP00000004945 | 93.3121 | 89.4904 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.0107 | 88.4409 | tetraodon_nigroviridis | ENSTNIP00000021832 | 93.0107 | 88.4409 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.2796 | 87.9032 | paramormyrops_kingsleyae | ENSPKIP00000004073 | 81.8396 | 77.1226 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.5484 | 88.9785 | scleropages_formosus | ENSSFOP00015022593 | 88.5496 | 84.2239 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.2796 | 88.4409 | leptobrachium_leishanense | ENSLLEP00000020315 | 92.0424 | 87.2679 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 93.5484 | 88.9785 | scophthalmus_maximus | ENSSMAP00000011182 | 88.5496 | 84.2239 |
| ENSG00000136807 | homo_sapiens | ENSP00000362361 | 84.6774 | 81.1828 | oryzias_melastigma | ENSOMEP00000003820 | 91.8367 | 88.0466 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0000978 | enables RNA polymerase II cis-regulatory region sequence-specific DNA binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0001223 | enables transcription coactivator binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0003677 | enables DNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0003682 | enables chromatin binding | 1 | ISS | Homo_sapiens(9606) | Function |
| GO:0004672 | enables protein kinase activity | 1 | TAS | Homo_sapiens(9606) | Function |
| GO:0004674 | enables protein serine/threonine kinase activity | 2 | IDA,TAS | Homo_sapiens(9606) | Function |
| GO:0004693 | enables cyclin-dependent protein serine/threonine kinase activity | 2 | IBA,IDA | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005524 | enables ATP binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0005634 | is_active_in nucleus | 3 | IBA,IDA | Homo_sapiens(9606) | Component |
| GO:0005654 | located_in nucleoplasm | 2 | IDA,TAS | Homo_sapiens(9606) | Component |
| GO:0006281 | involved_in DNA repair | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0006282 | involved_in regulation of DNA repair | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0006366 | involved_in transcription by RNA polymerase II | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0006367 | involved_in transcription initiation at RNA polymerase II promoter | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0006368 | involved_in transcription elongation by RNA polymerase II | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0006468 | involved_in protein phosphorylation | 3 | IBA,IDA | Homo_sapiens(9606) | Process |
| GO:0007346 | NOT involved_in regulation of mitotic cell cycle | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0008023 | part_of transcription elongation factor complex | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0008024 | part_of cyclin/CDK positive transcription elongation factor complex | 2 | IDA,IPI | Homo_sapiens(9606) | Component |
| GO:0008283 | involved_in cell population proliferation | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0008353 | enables RNA polymerase II CTD heptapeptide repeat kinase activity | 2 | IBA,IDA | Homo_sapiens(9606) | Function |
| GO:0016020 | located_in membrane | 1 | HDA | Homo_sapiens(9606) | Component |
| GO:0016301 | enables kinase activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0016605 | located_in PML body | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0019901 | enables protein kinase binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0031297 | involved_in replication fork processing | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0031440 | involved_in regulation of mRNA 3-end processing | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0032968 | involved_in positive regulation of transcription elongation by RNA polymerase II | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0036464 | located_in cytoplasmic ribonucleoprotein granule | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0043923 | involved_in positive regulation by host of viral transcription | 2 | IDA,ISS | Homo_sapiens(9606) | Process |
| GO:0045944 | involved_in positive regulation of transcription by RNA polymerase II | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0051147 | involved_in regulation of muscle cell differentiation | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0051647 | involved_in nucleus localization | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0051726 | involved_in regulation of cell cycle | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0070691 | part_of P-TEFb complex | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0071345 | involved_in cellular response to cytokine stimulus | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0097322 | enables 7SK snRNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0106310 | enables protein serine kinase activity | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0120186 | involved_in negative regulation of protein localization to chromatin | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0120187 | involved_in positive regulation of protein localization to chromatin | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0140673 | involved_in transcription elongation-coupled chromatin remodeling | 1 | IDA | Homo_sapiens(9606) | Process |