Welcome to
RBP World!
Functions and Diseases
| RBP Type | Canonical_RBPs |
| Diseases | CancerSARS-COV-OC43FlavivirusesHIVRVSARS-COV-2SINVDengueZika |
| Drug | N.A. |
| Main interacting RNAs | mRNA |
| Moonlighting functions | N.A. |
| Localizations | Stress granucleP-body |
| BulkPerturb-seq | DataSet_01_111 DataSet_01_270 DataSet_01_362 DataSet_02_18 |
Description
| Ensembl ID | ENSG00000136231 | Gene ID | 10643 | Accession | 28868 |
| Symbol | IGF2BP3 | Alias | KOC;CT98;IMP3;KOC1;IMP-3;VICKZ3 | Full Name | insulin like growth factor 2 mRNA binding protein 3 |
| Status | Confidence | Length | 160283 bases | Strand | Minus strand |
| Position | 7 : 23310209 - 23470491 | RNA binding domain | KH_1 , RRM_1 | ||
| Summary | The protein encoded by this gene is primarily found in the nucleolus, where it can bind to the 5' UTR of the insulin-like growth factor II leader 3 mRNA and may repress translation of insulin-like growth factor II during late development. The encoded protein contains several KH domains, which are important in RNA binding and are known to be involved in RNA synthesis and metabolism. A pseudogene exists on chromosome 7, and there are putative pseudogenes on other chromosomes. [provided by RefSeq, Jul 2008] | ||||
RNA binding domains (RBDs)
| Protein | Domain | Pfam ID | E-value | Domain number |
|---|---|---|---|---|
| ENSP00000258729 | RRM_1 | PF00076 | 2.3e-17 | 1 |
| ENSP00000258729 | RRM_1 | PF00076 | 2.3e-17 | 2 |
| ENSP00000258729 | KH_1 | PF00013 | 2.3e-56 | 3 |
| ENSP00000258729 | KH_1 | PF00013 | 2.3e-56 | 4 |
| ENSP00000258729 | KH_1 | PF00013 | 2.3e-56 | 5 |
| ENSP00000258729 | KH_1 | PF00013 | 2.3e-56 | 6 |
| ENSP00000395936 | RRM_1 | PF00076 | 1.6e-07 | 1 |
| ENSP00000480267 | KH_1 | PF00013 | 1.2e-27 | 1 |
| ENSP00000480267 | KH_1 | PF00013 | 1.2e-27 | 2 |
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|---|---|---|---|
| 37305324 | IGF2BP3-induced activation of EIF5B contributes to progression of hepatocellular carcinoma cells. | Xiaoyin Li | 2022-01-01 | Oncology research |
| 32083017 | Overexpression of IGF2BP3 as a Potential Oncogene in Ovarian Clear Cell Carcinoma. | Huidi Liu | 2019-01-01 | Frontiers in oncology |
| 35676262 | IGF2BP3 enhances the mRNA stability of E2F3 by interacting with LINC00958 to promote endometrial carcinoma progression. | Cuicui Wang | 2022-06-08 | Cell death discovery |
| 33805930 | IGF2BP3 Associates with Proliferative Phenotype and Prognostic Features in B-Cell Acute Lymphoblastic Leukemia. | Artturi Mäkinen | 2021-03-25 | Cancers |
| 34986849 | Hsa_circ_0003258 promotes prostate cancer metastasis by complexing with IGF2BP3 and sponging miR-653-5p. | Yu-Zhong Yu | 2022-01-05 | Molecular cancer |
| 30760837 | DNA demethylation is associated with malignant progression of lower-grade gliomas. | Masashi Nomura | 2019-02-13 | Scientific reports |
| 31118463 | Combinatorial recognition of clustered RNA elements by the multidomain RNA-binding protein IMP3. | Tim Schneider | 2019-05-22 | Nature communications |
| 22840368 | Insulin-like growth factor 2 messenger RNA binding protein 3 (IGF2BP3) is a marker of unfavourable prognosis in colorectal cancer. | Paul Lochhead | 2012-12-01 | European journal of cancer (Oxford, England : 1990) |
| 37207381 | Advances in lung adenocarcinoma: A novel perspective on prognoses and immune responses of CENPO as an oncogenic superenhancer. | Tongdong Shi | 2023-08-01 | Translational oncology |
| 33279839 | IGF2BP3 Expression Correlates With Poor Prognosis in Esophageal Squamous Cell Carcinoma. | Akiyuki Wakita | 2021-03-01 | The Journal of surgical research |
| 34008756 | Prognostic implications of immune-related eight-gene signature in pediatric brain tumors. | Yi Wang | 2021-01-01 | Brazilian journal of medical and biological research = Revista brasileira de pesquisas medicas e biologicas |
| 36831493 | RNA-Binding Proteins in Bladder Cancer. | Yuanhui Gao | 2023-02-10 | Cancers |
| 36761737 | Multi-omics analysis of N6-methyladenosine reader IGF2BP3 as a promising biomarker in pan-cancer. | Pin Chen | 2023-01-01 | Frontiers in immunology |
| 36030651 | IGF2BP3-NRF2 axis regulates ferroptosis in hepatocellular carcinoma. | Zhihua Lu | 2022-10-30 | Biochemical and biophysical research communications |
| 29186811 | Ascorbic Acid Attenuates Senescence of Human Osteoarthritic Osteoblasts. | G Maximilian Burger | 2017-11-24 | International journal of molecular sciences |
| 37407973 | CircGNB1 facilitates the malignant phenotype of GSCs by regulating miR-515-5p/miR-582-3p-XPR1 axis. | Jinpeng Hu | 2023-07-05 | Cancer cell international |
| 25216519 | IGF2BP3-mediated translation in cell protrusions promotes cell invasiveness and metastasis of pancreatic cancer. | Keisuke Taniuchi | 2014-08-30 | Oncotarget |
| 36338681 | Polymorphic variants of IGF2BP3 and SENCR have an impact on predisposition and/or progression of Ewing sarcoma. | Marcella Martinelli | 2022-01-01 | Frontiers in oncology |
| 31451669 | Mechanistic basis of neonatal heart regeneration revealed by transcriptome and histone modification profiling. | Zhaoning Wang | 2019-09-10 | Proceedings of the National Academy of Sciences of the United States of America |
| 37353558 | Extracellular vesicle-associated IGF2BP3 tunes Ewing sarcoma cell migration and affects PI3K/Akt pathway in neighboring cells. | Caterina Mancarella | 2023-06-23 | Cancer gene therapy |
| 27941992 | High Expression of KIF20A Is Associated with Poor Overall Survival and Tumor Progression in Early-Stage Cervical Squamous Cell Carcinoma. | Weijing Zhang | 2016-01-01 | PloS one |
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| IGF2BP3-201 | ENST00000258729 | 4274 | 579aa | ENSP00000258729 |
| IGF2BP3-202 | ENST00000421467 | 2988 | 81aa | ENSP00000395936 |
| IGF2BP3-217 | ENST00000498363 | 529 | No protein | - |
| IGF2BP3-205 | ENST00000467592 | 600 | No protein | - |
| IGF2BP3-216 | ENST00000497563 | 657 | No protein | - |
| IGF2BP3-212 | ENST00000480547 | 596 | No protein | - |
| IGF2BP3-215 | ENST00000495009 | 452 | No protein | - |
| IGF2BP3-207 | ENST00000468263 | 706 | No protein | - |
| IGF2BP3-214 | ENST00000492771 | 564 | No protein | - |
| IGF2BP3-211 | ENST00000479504 | 559 | No protein | - |
| IGF2BP3-204 | ENST00000466809 | 588 | No protein | - |
| IGF2BP3-213 | ENST00000491719 | 800 | No protein | - |
| IGF2BP3-203 | ENST00000465058 | 534 | No protein | - |
| IGF2BP3-208 | ENST00000469723 | 556 | No protein | - |
| IGF2BP3-209 | ENST00000474105 | 663 | No protein | - |
| IGF2BP3-210 | ENST00000476938 | 500 | No protein | - |
| IGF2BP3-206 | ENST00000468005 | 2948 | No protein | - |
| IGF2BP3-218 | ENST00000619562 | 2126 | 198aa | ENSP00000480267 |
Phenotypes
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| 32731 | Body Mass Index | 4.5725990E-005 | - |
| 32817 | Body Mass Index | 5.0777281E-005 | - |
| 63249 | Body Height | 2E-14 | 20881960 |
| 75104 | Hip | 2E-8 | 25673412 |
| 75105 | Body Mass Index | 2E-8 | 25673412 |
| 85544 | Body Height | 4E-26 | 25282103 |
| 85944 | Body Height | 5E-10 | 23563607 |
| 86008 | Personality | 5E-6 | 23903073 |
| 87462 | Hip | 1E-8 | 25673412 |
| 87463 | Body Mass Index | 1E-8 | 25673412 |
| 88388 | Body Height | 8E-8 | 25281659 |
| 94137 | Immunoglobulins | 2E-6 | 23382691 |
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
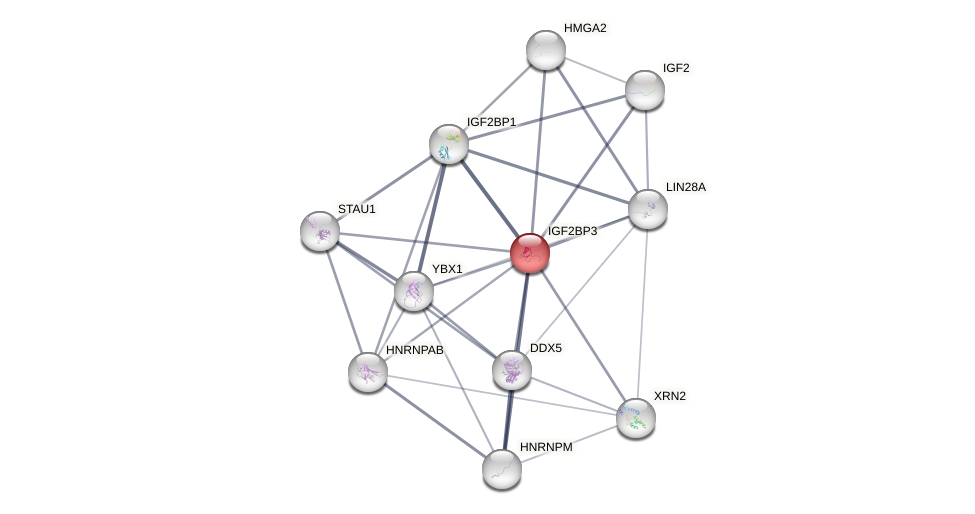
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000136231 | homo_sapiens | ENSP00000290341 | 83.5355 | 73.3102 | homo_sapiens | ENSP00000258729 | 83.247 | 73.057 |
| ENSG00000136231 | homo_sapiens | ENSP00000417196 | 37.2208 | 20.0993 | homo_sapiens | ENSP00000258729 | 25.9067 | 13.9896 |
| ENSG00000136231 | homo_sapiens | ENSP00000263257 | 26.8293 | 13.8211 | homo_sapiens | ENSP00000258729 | 22.7979 | 11.7444 |
| ENSG00000136231 | homo_sapiens | ENSP00000505796 | 39.8922 | 22.9111 | homo_sapiens | ENSP00000258729 | 25.5613 | 14.6805 |
| ENSG00000136231 | homo_sapiens | ENSP00000365439 | 31.0345 | 17.6724 | homo_sapiens | ENSP00000258729 | 24.8705 | 14.1623 |
| ENSG00000136231 | homo_sapiens | ENSP00000448762 | 41.9889 | 24.5856 | homo_sapiens | ENSP00000258729 | 26.2522 | 15.3713 |
| ENSG00000136231 | homo_sapiens | ENSP00000371634 | 76.7947 | 63.606 | homo_sapiens | ENSP00000258729 | 79.4473 | 65.8031 |
| ENSG00000136231 | homo_sapiens | ENSP00000359804 | 29.8137 | 16.9255 | homo_sapiens | ENSP00000258729 | 33.1606 | 18.8256 |
| ENSG00000136231 | homo_sapiens | ENSP00000318177 | 32.6923 | 18.007 | homo_sapiens | ENSP00000258729 | 32.2971 | 17.7893 |
| ENSG00000136231 | homo_sapiens | ENSP00000305556 | 41.8539 | 24.1573 | homo_sapiens | ENSP00000258729 | 25.734 | 14.8532 |
| ENSG00000136231 | homo_sapiens | ENSP00000438875 | 26.2327 | 13.8067 | homo_sapiens | ENSP00000258729 | 22.9706 | 12.0898 |
| ENSG00000136231 | homo_sapiens | ENSP00000471146 | 27.577 | 15.7965 | homo_sapiens | ENSP00000258729 | 35.5786 | 20.38 |
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 99.4819 | 99.1364 | nomascus_leucogenys | ENSNLEP00000017985 | 98.2935 | 97.9522 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 100 | 100 | pan_paniscus | ENSPPAP00000021170 | 100 | 100 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 100 | 100 | pongo_abelii | ENSPPYP00000042932 | 100 | 100 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 100 | 100 | pan_troglodytes | ENSPTRP00000032446 | 100 | 100 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 89.81 | 89.81 | gorilla_gorilla | ENSGGOP00000020311 | 100 | 100 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 92.7461 | 86.3558 | ornithorhynchus_anatinus | ENSOANP00000036315 | 89.6494 | 83.4725 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 83.7651 | 74.6114 | sarcophilus_harrisii | ENSSHAP00000008269 | 67.4548 | 60.0835 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 68.7392 | 61.4853 | notamacropus_eugenii | ENSMEUP00000011416 | 68.3849 | 61.1684 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 89.9827 | 89.1192 | choloepus_hoffmanni | ENSCHOP00000002070 | 89.9827 | 89.1192 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 86.0104 | 84.6287 | dasypus_novemcinctus | ENSDNOP00000030269 | 90.2174 | 88.7681 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 69.4301 | 68.9119 | erinaceus_europaeus | ENSEEUP00000004308 | 69.4301 | 68.9119 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 83.247 | 82.038 | echinops_telfairi | ENSETEP00000012623 | 92.1606 | 90.8222 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 99.4819 | 99.3092 | callithrix_jacchus | ENSCJAP00000002684 | 99.4819 | 99.3092 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 100 | 99.8273 | cercocebus_atys | ENSCATP00000025047 | 100 | 99.8273 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 100 | 99.8273 | macaca_fascicularis | ENSMFAP00000032754 | 94.918 | 94.7541 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 100 | 99.8273 | macaca_mulatta | ENSMMUP00000035154 | 100 | 99.8273 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 100 | 99.8273 | macaca_nemestrina | ENSMNEP00000010144 | 100 | 99.8273 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 100 | 99.8273 | papio_anubis | ENSPANP00000007781 | 100 | 99.8273 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 100 | 99.8273 | mandrillus_leucophaeus | ENSMLEP00000000835 | 100 | 99.8273 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 99.3092 | 98.6183 | tursiops_truncatus | ENSTTRP00000008462 | 99.3092 | 98.6183 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 99.1364 | 98.6183 | vulpes_vulpes | ENSVVUP00000001267 | 99.1364 | 98.6183 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 98.2729 | 96.5458 | mus_spretus | MGP_SPRETEiJ_P0076619 | 98.2729 | 96.5458 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 98.9637 | 98.1002 | equus_asinus | ENSEASP00005059416 | 98.9637 | 98.1002 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 98.791 | 97.582 | capra_hircus | ENSCHIP00000031520 | 98.6207 | 97.4138 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 83.0743 | 82.7288 | loxodonta_africana | ENSLAFP00000024507 | 98.5656 | 98.1557 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 99.3092 | 98.791 | panthera_leo | ENSPLOP00000008490 | 99.3092 | 98.791 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 99.1364 | 98.2729 | ailuropoda_melanoleuca | ENSAMEP00000003676 | 99.1364 | 98.2729 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 99.1364 | 97.7548 | bos_indicus_hybrid | ENSBIXP00005017074 | 98.9655 | 97.5862 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 88.4283 | 87.3921 | oryctolagus_cuniculus | ENSOCUP00000007754 | 97.1537 | 96.0152 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 34.1969 | 34.1969 | equus_caballus | ENSECAP00000014668 | 100 | 100 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 16.9257 | 14.1623 | equus_caballus | ENSECAP00000058014 | 83.0508 | 69.4915 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 99.3092 | 98.6183 | canis_lupus_familiaris | ENSCAFP00845012868 | 99.3092 | 98.6183 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 99.3092 | 98.6183 | panthera_pardus | ENSPPRP00000026934 | 99.3092 | 98.6183 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 99.3092 | 98.9637 | marmota_marmota_marmota | ENSMMMP00000005912 | 99.3092 | 98.9637 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 99.1364 | 98.4456 | balaenoptera_musculus | ENSBMSP00010010072 | 99.1364 | 98.4456 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 88.9465 | 88.2556 | cavia_porcellus | ENSCPOP00000013584 | 99.2293 | 98.4586 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 99.3092 | 98.6183 | felis_catus | ENSFCAP00000016657 | 99.3092 | 98.6183 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 81.3472 | 80.829 | ursus_americanus | ENSUAMP00000028171 | 96.7146 | 96.0986 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 87.7375 | 79.4473 | monodelphis_domestica | ENSMODP00000001219 | 83.4154 | 75.5337 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 97.2366 | 95.8549 | bison_bison_bison | ENSBBBP00000022270 | 98.9455 | 97.5395 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 99.3092 | 98.6183 | monodon_monoceros | ENSMMNP00015018542 | 99.3092 | 98.6183 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 88.7738 | 86.7012 | sus_scrofa | ENSSSCP00000044238 | 87.7133 | 85.6655 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 99.6546 | 99.4819 | microcebus_murinus | ENSMICP00000012488 | 99.6546 | 99.4819 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 15.0259 | 13.2988 | octodon_degus | ENSODEP00000017576 | 61.7021 | 54.6099 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 12.7807 | 12.4352 | octodon_degus | ENSODEP00000012243 | 93.6709 | 91.1392 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 98.4456 | 97.582 | jaculus_jaculus | ENSJJAP00000006521 | 98.4456 | 97.582 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 98.791 | 97.582 | ovis_aries_rambouillet | ENSOARP00020008848 | 98.791 | 97.582 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 94.4732 | 92.4007 | ursus_maritimus | ENSUMAP00000001114 | 95.6294 | 93.5315 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 71.5026 | 70.2936 | ochotona_princeps | ENSOPRP00000009595 | 82.8 | 81.4 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 99.3092 | 98.6183 | delphinapterus_leucas | ENSDLEP00000030523 | 99.3092 | 98.6183 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 99.3092 | 98.6183 | physeter_catodon | ENSPCTP00005004454 | 99.3092 | 98.6183 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 98.2729 | 96.7185 | mus_caroli | MGP_CAROLIEiJ_P0073751 | 98.2729 | 96.7185 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 98.2729 | 96.5458 | mus_musculus | ENSMUSP00000031838 | 98.2729 | 96.5458 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 98.9637 | 98.2729 | dipodomys_ordii | ENSDORP00000013339 | 98.9637 | 98.2729 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 98.9637 | 98.1002 | mustela_putorius_furo | ENSMPUP00000013424 | 98.9637 | 98.1002 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 99.4819 | 98.791 | aotus_nancymaae | ENSANAP00000008062 | 99.4819 | 98.791 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 99.4819 | 99.3092 | saimiri_boliviensis_boliviensis | ENSSBOP00000005460 | 99.4819 | 99.3092 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 89.2919 | 89.1192 | vicugna_pacos | ENSVPAP00000000568 | 99.4231 | 99.2308 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 83.4197 | 83.4197 | chlorocebus_sabaeus | ENSCSAP00000010118 | 99.7934 | 99.7934 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 71.3299 | 70.639 | tupaia_belangeri | ENSTBEP00000001016 | 79.2706 | 78.5029 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 97.2366 | 95.6822 | mesocricetus_auratus | ENSMAUP00000002294 | 97.2366 | 95.6822 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 99.1364 | 98.4456 | phocoena_sinus | ENSPSNP00000013365 | 99.1364 | 98.4456 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 99.3092 | 98.9637 | camelus_dromedarius | ENSCDRP00005000200 | 99.3092 | 98.9637 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 99.6546 | 99.1364 | carlito_syrichta | ENSTSYP00000011618 | 99.6546 | 99.1364 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 99.3092 | 98.6183 | panthera_tigris_altaica | ENSPTIP00000003215 | 99.3092 | 98.6183 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 98.2729 | 96.8912 | rattus_norvegicus | ENSRNOP00000071626 | 98.2729 | 96.8912 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 87.0466 | 85.8377 | prolemur_simus | ENSPSMP00000006045 | 94.7368 | 93.4211 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 96.8912 | 95.3368 | microtus_ochrogaster | ENSMOCP00000002216 | 96.8912 | 95.3368 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 80.829 | 80.1382 | sorex_araneus | ENSSARP00000005970 | 89.313 | 88.5496 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 97.582 | 95.8549 | peromyscus_maniculatus_bairdii | ENSPEMP00000027724 | 97.582 | 95.8549 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 99.1364 | 97.7548 | bos_mutus | ENSBMUP00000029301 | 99.1364 | 97.7548 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 98.2729 | 96.5458 | mus_spicilegus | ENSMSIP00000024188 | 98.2729 | 96.5458 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 97.582 | 95.6822 | cricetulus_griseus_chok1gshd | ENSCGRP00001009789 | 97.582 | 95.6822 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 79.2746 | 78.7565 | pteropus_vampyrus | ENSPVAP00000010648 | 88.2692 | 87.6923 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 82.9016 | 81.8653 | myotis_lucifugus | ENSMLUP00000007711 | 94.4882 | 93.3071 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 82.7288 | 73.9206 | vombatus_ursinus | ENSVURP00010026257 | 83.7413 | 74.8252 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 82.7288 | 73.9206 | vombatus_ursinus | ENSVURP00010002118 | 83.7413 | 74.8252 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 99.1364 | 98.791 | rhinolophus_ferrumequinum | ENSRFEP00010013955 | 99.1364 | 98.791 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 95.1641 | 93.6097 | neovison_vison | ENSNVIP00000016663 | 91.9866 | 90.4841 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 99.1364 | 97.7548 | bos_taurus | ENSBTAP00000025854 | 98.9655 | 97.5862 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 99.3092 | 98.2729 | heterocephalus_glaber_female | ENSHGLP00000018267 | 99.3092 | 98.2729 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 100 | 99.8273 | rhinopithecus_bieti | ENSRBIP00000018039 | 100 | 99.8273 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 99.3092 | 98.6183 | canis_lupus_dingo | ENSCAFP00020003212 | 99.3092 | 98.6183 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 13.6442 | 13.4715 | sciurus_vulgaris | ENSSVLP00005024388 | 100 | 98.7342 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 34.0242 | 33.8515 | sciurus_vulgaris | ENSSVLP00005024271 | 100 | 99.4924 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 87.7375 | 78.5838 | phascolarctos_cinereus | ENSPCIP00000009727 | 80.1262 | 71.7666 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 63.9033 | 63.0397 | procavia_capensis | ENSPCAP00000009425 | 71.8447 | 70.8738 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 98.6183 | 97.9275 | nannospalax_galili | ENSNGAP00000025715 | 98.6183 | 97.9275 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 99.1364 | 97.7548 | bos_grunniens | ENSBGRP00000000500 | 95.1907 | 93.864 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 99.1364 | 98.791 | chinchilla_lanigera | ENSCLAP00000014253 | 99.1364 | 98.791 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 98.6183 | 97.0639 | cervus_hanglu_yarkandensis | ENSCHYP00000020352 | 93.7603 | 92.2824 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 97.582 | 96.8912 | catagonus_wagneri | ENSCWAP00000001501 | 97.582 | 96.8912 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 99.3092 | 98.9637 | ictidomys_tridecemlineatus | ENSSTOP00000007642 | 99.3092 | 98.9637 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 99.6546 | 99.6546 | otolemur_garnettii | ENSOGAP00000015371 | 99.6546 | 99.6546 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 99.4819 | 99.3092 | cebus_imitator | ENSCCAP00000019710 | 99.4819 | 99.3092 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 99.3092 | 98.9637 | urocitellus_parryii | ENSUPAP00010019165 | 99.3092 | 98.9637 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 98.9637 | 97.582 | moschus_moschiferus | ENSMMSP00000033135 | 98.9637 | 97.582 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 100 | 99.8273 | rhinopithecus_roxellana | ENSRROP00000023027 | 100 | 99.8273 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 98.2729 | 96.8912 | mus_pahari | MGP_PahariEiJ_P0049842 | 98.2729 | 96.8912 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 13.6442 | 13.4715 | propithecus_coquereli | ENSPCOP00000010880 | 100 | 98.7342 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 19.3437 | 8.80829 | caenorhabditis_elegans | Y119D3B.17.2 | 25.9259 | 11.8056 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 15.0259 | 7.94473 | caenorhabditis_elegans | Y59A8B.10d.1 | 20.8633 | 11.0312 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 25.9067 | 14.1623 | caenorhabditis_elegans | C36E6.1b.1 | 23.4009 | 12.7925 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 25.5613 | 12.9534 | caenorhabditis_elegans | M01A10.1.1 | 25.256 | 12.7986 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 43.3506 | 25.5613 | caenorhabditis_elegans | M88.5c.1 | 29.3911 | 17.3302 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 26.0794 | 13.9896 | drosophila_melanogaster | FBpp0293521 | 26.3525 | 14.1361 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 22.4525 | 10.7081 | drosophila_melanogaster | FBpp0306419 | 16.6667 | 7.94872 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 51.2953 | 35.4059 | drosophila_melanogaster | FBpp0293680 | 46.5517 | 32.1317 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 20.7254 | 11.0535 | ciona_intestinalis | ENSCINP00000001984 | 33.6134 | 17.9272 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 17.7893 | 8.46287 | ciona_intestinalis | ENSCINP00000025950 | 36.2676 | 17.2535 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 62.6943 | 45.4231 | ciona_intestinalis | ENSCINP00000008726 | 61.4213 | 44.5008 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 62.3489 | 54.4041 | petromyzon_marinus | ENSPMAP00000003549 | 78.4783 | 68.4783 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 25.734 | 19.171 | eptatretus_burgeri | ENSEBUP00000009195 | 71.6346 | 53.3654 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 80.3109 | 72.1934 | callorhinchus_milii | ENSCMIP00000040392 | 83.6331 | 75.1799 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 69.4301 | 63.3851 | latimeria_chalumnae | ENSLACP00000014733 | 83.5759 | 76.2994 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 88.7738 | 80.4836 | lepisosteus_oculatus | ENSLOCP00000014265 | 83.3063 | 75.5267 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 85.1468 | 74.7841 | clupea_harengus | ENSCHAP00000029015 | 82.3038 | 72.2871 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 86.0104 | 77.0294 | danio_rerio | ENSDARP00000010878 | 85.567 | 76.6323 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 86.1831 | 76.8567 | carassius_auratus | ENSCARP00000066873 | 83.1667 | 74.1667 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 83.0743 | 74.266 | carassius_auratus | ENSCARP00000047141 | 76.96 | 68.8 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 86.0104 | 77.0294 | astyanax_mexicanus | ENSAMXP00000043052 | 81.7734 | 73.2348 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 84.6287 | 75.6477 | ictalurus_punctatus | ENSIPUP00000008925 | 84.4828 | 75.5172 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 85.6649 | 76.3385 | esox_lucius | ENSELUP00000073510 | 75.4947 | 67.2755 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 84.8014 | 74.7841 | electrophorus_electricus | ENSEEEP00000017253 | 81.157 | 71.5703 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 85.4922 | 76.1658 | oncorhynchus_kisutch | ENSOKIP00005100849 | 79.3269 | 70.6731 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 84.9741 | 75.3022 | oncorhynchus_kisutch | ENSOKIP00005013884 | 76.7551 | 68.0187 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 85.4922 | 76.1658 | oncorhynchus_mykiss | ENSOMYP00000028030 | 79.3269 | 70.6731 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 84.9741 | 75.3022 | oncorhynchus_mykiss | ENSOMYP00000015341 | 78.2194 | 69.3164 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 85.4922 | 76.1658 | salmo_salar | ENSSSAP00000033375 | 78.9474 | 70.3349 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 84.9741 | 75.3022 | salmo_salar | ENSSSAP00000033367 | 78.9727 | 69.9839 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 84.9741 | 75.475 | salmo_trutta | ENSSTUP00000104972 | 78.2194 | 69.4754 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 85.3195 | 76.1658 | salmo_trutta | ENSSTUP00000047011 | 79.04 | 70.56 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 80.829 | 70.9845 | gadus_morhua | ENSGMOP00000037439 | 75.4839 | 66.2903 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 84.456 | 74.7841 | fundulus_heteroclitus | ENSFHEP00000014331 | 83.3049 | 73.7649 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 84.8014 | 74.9568 | poecilia_reticulata | ENSPREP00000002455 | 80.7566 | 71.3816 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 84.8014 | 75.3022 | xiphophorus_maculatus | ENSXMAP00000028569 | 80.7566 | 71.7105 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 84.6287 | 75.6477 | oryzias_latipes | ENSORLP00000005931 | 80.4598 | 71.9212 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 83.5924 | 74.4387 | cyclopterus_lumpus | ENSCLMP00005041067 | 80.3987 | 71.5947 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 81.0017 | 71.5026 | oreochromis_niloticus | ENSONIP00000045474 | 76.509 | 67.5367 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 84.8014 | 75.8204 | haplochromis_burtoni | ENSHBUP00000016232 | 83.3616 | 74.5331 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 84.8014 | 75.8204 | astatotilapia_calliptera | ENSACLP00000000064 | 83.5034 | 74.6599 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 83.7651 | 73.9206 | sparus_aurata | ENSSAUP00010053601 | 80.1653 | 70.7438 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 79.9655 | 71.5026 | lates_calcarifer | ENSLCAP00010036454 | 79.2808 | 70.8904 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 84.1105 | 77.0294 | xenopus_tropicalis | ENSXETP00000016106 | 87.4327 | 80.0718 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 92.228 | 85.3195 | chrysemys_picta_bellii | ENSCPBP00000032001 | 91.4384 | 84.589 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 88.2556 | 81.3472 | sphenodon_punctatus | ENSSPUP00000007974 | 89.8067 | 82.7768 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 89.2919 | 82.3834 | crocodylus_porosus | ENSCPRP00005022919 | 82.4561 | 76.0766 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 48.532 | 44.3869 | laticauda_laticaudata | ENSLLTP00000008356 | 88.6435 | 81.0726 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 12.6079 | 11.399 | notechis_scutatus | ENSNSUP00000020877 | 85.8824 | 77.6471 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 45.5959 | 42.6598 | notechis_scutatus | ENSNSUP00000020878 | 92.6316 | 86.6667 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 87.3921 | 80.3109 | pseudonaja_textilis | ENSPTXP00000020532 | 89.8757 | 82.5933 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 92.228 | 86.0104 | anas_platyrhynchos_platyrhynchos | ENSAPLP00000013116 | 91.4384 | 85.274 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 89.9827 | 83.7651 | gallus_gallus | ENSGALP00010001856 | 83.2268 | 77.476 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 82.2107 | 75.6477 | meleagris_gallopavo | ENSMGAP00000026063 | 88.1481 | 81.1111 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 82.3834 | 75.8204 | serinus_canaria | ENSSCAP00000020687 | 83.101 | 76.4808 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 92.228 | 86.0104 | parus_major | ENSPMJP00000000881 | 91.4384 | 85.274 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 92.0553 | 84.8014 | anolis_carolinensis | ENSACAP00000013341 | 91.2671 | 84.0753 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 76.1658 | 67.7029 | neolamprologus_brichardi | ENSNBRP00000007654 | 83.5227 | 74.2424 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 87.5648 | 80.3109 | naja_naja | ENSNNAP00000020829 | 85.2101 | 78.1513 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 86.7012 | 79.4473 | podarcis_muralis | ENSPMRP00000026962 | 72.2302 | 66.187 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 81.6926 | 75.3022 | geospiza_fortis | ENSGFOP00000015202 | 84.767 | 78.1362 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 92.228 | 86.0104 | taeniopygia_guttata | ENSTGUP00000002872 | 91.4384 | 85.274 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 87.2193 | 79.4473 | erpetoichthys_calabaricus | ENSECRP00000017028 | 88.4413 | 80.5604 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 83.9378 | 74.9568 | kryptolebias_marmoratus | ENSKMAP00000026764 | 80.4636 | 71.8543 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 55.6131 | 44.905 | salvator_merianae | ENSSMRP00000026847 | 80.7018 | 65.1629 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 91.7098 | 84.1105 | salvator_merianae | ENSSMRP00000004141 | 86.3415 | 79.187 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 77.0294 | 67.3575 | oryzias_javanicus | ENSOJAP00000012872 | 78.3831 | 68.5413 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 83.5924 | 74.7841 | amphilophus_citrinellus | ENSACIP00000001728 | 83.1615 | 74.3986 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 84.456 | 75.8204 | dicentrarchus_labrax | ENSDLAP00005016844 | 80.6931 | 72.4422 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 84.1105 | 75.475 | amphiprion_percula | ENSAPEP00000026206 | 83.677 | 75.0859 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 84.6287 | 75.475 | oryzias_sinensis | ENSOSIP00000020990 | 80.5921 | 71.875 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 91.3644 | 83.7651 | pelodiscus_sinensis | ENSPSIP00000015996 | 90.4274 | 82.906 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 88.601 | 81.6926 | gopherus_evgoodei | ENSGEVP00005026311 | 91.771 | 84.6154 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 92.228 | 85.1468 | chelonoidis_abingdonii | ENSCABP00000000679 | 91.2821 | 84.2735 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 84.1105 | 75.475 | seriola_dumerili | ENSSDUP00000007583 | 83.677 | 75.0859 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 83.0743 | 74.0933 | cottoperca_gobio | ENSCGOP00000009974 | 79.7678 | 71.1443 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 79.7927 | 72.5389 | struthio_camelus_australis | ENSSCUP00000009675 | 82.5 | 75 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 81.1744 | 73.5751 | anser_brachyrhynchus | ENSABRP00000011755 | 77.9436 | 70.6468 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 69.0846 | 61.4853 | gasterosteus_aculeatus | ENSGACP00000009196 | 75.6144 | 67.2968 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 81.0017 | 70.8117 | cynoglossus_semilaevis | ENSCSEP00000018959 | 78.6913 | 68.7919 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 84.9741 | 75.1295 | poecilia_formosa | ENSPFOP00000006186 | 80.7882 | 71.4286 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 83.247 | 74.0933 | myripristis_murdjan | ENSMMDP00005047875 | 81.5567 | 72.5888 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 83.0743 | 73.9206 | sander_lucioperca | ENSSLUP00000004135 | 80.3005 | 71.4524 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 92.228 | 86.0104 | strigops_habroptila | ENSSHBP00005006922 | 91.4384 | 85.274 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 11.7444 | 10.0173 | amphiprion_ocellaris | ENSAOCP00000026917 | 83.9506 | 71.6049 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 64.2487 | 58.3765 | amphiprion_ocellaris | ENSAOCP00000011225 | 83.2215 | 75.6152 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 92.228 | 85.3195 | terrapene_carolina_triunguis | ENSTMTP00000006916 | 91.4384 | 84.589 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 85.1468 | 75.6477 | hucho_hucho | ENSHHUP00000070865 | 78.3784 | 69.6343 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 85.4922 | 76.1658 | hucho_hucho | ENSHHUP00000007018 | 78.6963 | 70.1113 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 84.6287 | 74.9568 | betta_splendens | ENSBSLP00000032793 | 80.7249 | 71.4992 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 85.4922 | 77.0294 | pygocentrus_nattereri | ENSPNAP00000000882 | 84.9057 | 76.5009 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 84.456 | 75.3022 | anabas_testudineus | ENSATEP00000000999 | 80.8264 | 72.0661 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 17.2712 | 9.32642 | ciona_savignyi | ENSCSAVP00000004755 | 47.619 | 25.7143 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 9.67185 | 4.66321 | ciona_savignyi | ENSCSAVP00000007214 | 45.9016 | 22.1311 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 62.1762 | 44.7323 | ciona_savignyi | ENSCSAVP00000018813 | 62.069 | 44.6552 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 84.1105 | 75.475 | seriola_lalandi_dorsalis | ENSSLDP00000033441 | 83.5334 | 74.9571 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 83.4197 | 73.7478 | labrus_bergylta | ENSLBEP00000006988 | 79.703 | 70.462 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 76.1658 | 67.7029 | pundamilia_nyererei | ENSPNYP00000026795 | 83.5227 | 74.2424 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 84.9741 | 75.8204 | mastacembelus_armatus | ENSMAMP00000065874 | 81.0544 | 72.3229 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 76.6839 | 66.494 | stegastes_partitus | ENSSPAP00000005376 | 80.1444 | 69.4946 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 85.4922 | 76.1658 | oncorhynchus_tshawytscha | ENSOTSP00005021982 | 79.3269 | 70.6731 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 84.9741 | 75.3022 | oncorhynchus_tshawytscha | ENSOTSP00005008093 | 78.2194 | 69.3164 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 84.2832 | 75.6477 | acanthochromis_polyacanthus | ENSAPOP00000030639 | 83.705 | 75.1286 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 92.228 | 86.0104 | aquila_chrysaetos_chrysaetos | ENSACCP00020012884 | 91.4384 | 85.274 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 84.8014 | 75.8204 | nothobranchius_furzeri | ENSNFUP00015040533 | 81.2914 | 72.6821 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 76.5112 | 67.3575 | cyprinodon_variegatus | ENSCVAP00000007245 | 82.6493 | 72.7612 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 81.1744 | 70.9845 | denticeps_clupeoides | ENSDCDP00000028304 | 81.1744 | 70.9845 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 82.5561 | 73.5751 | larimichthys_crocea | ENSLCRP00005038535 | 78.4893 | 69.9507 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 83.0743 | 73.5751 | takifugu_rubripes | ENSTRUP00000059529 | 79.5041 | 70.4132 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 90.3282 | 83.7651 | ficedula_albicollis | ENSFALP00000032097 | 88.9456 | 82.483 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 77.5475 | 68.7392 | hippocampus_comes | ENSHCOP00000020336 | 82.5368 | 73.1618 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 86.0104 | 77.0294 | cyprinus_carpio_carpio | ENSCCRP00000175808 | 82.7243 | 74.0864 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 85.8377 | 77.3748 | cyprinus_carpio_carpio | ENSCCRP00000076161 | 80.9446 | 72.9642 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 91.8826 | 85.3195 | coturnix_japonica | ENSCJPP00005015024 | 90.6303 | 84.1567 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 27.9793 | 25.2159 | poecilia_latipinna | ENSPLAP00000024073 | 78.6408 | 70.8738 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 84.9741 | 75.1295 | poecilia_latipinna | ENSPLAP00000005756 | 83.959 | 74.2321 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 85.8377 | 77.3748 | sinocyclocheilus_grahami | ENSSGRP00000103327 | 81.8781 | 73.8056 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 84.2832 | 75.475 | sinocyclocheilus_grahami | ENSSGRP00000097799 | 82.1549 | 73.569 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 84.8014 | 75.8204 | maylandia_zebra | ENSMZEP00005012220 | 83.3616 | 74.5331 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 82.7288 | 73.2297 | tetraodon_nigroviridis | ENSTNIP00000020906 | 82.3024 | 72.8522 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 84.456 | 75.3022 | paramormyrops_kingsleyae | ENSPKIP00000032155 | 82.8814 | 73.8983 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 86.7012 | 76.6839 | scleropages_formosus | ENSSFOP00015069962 | 83.8063 | 74.1235 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 91.3644 | 83.7651 | leptobrachium_leishanense | ENSLLEP00000009513 | 87.0066 | 79.7697 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 82.2107 | 72.8843 | scophthalmus_maximus | ENSSMAP00000023451 | 74.259 | 65.8346 |
| ENSG00000136231 | homo_sapiens | ENSP00000258729 | 84.2832 | 75.1295 | oryzias_melastigma | ENSOMEP00000016278 | 83.4188 | 74.359 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0000932 | located_in P-body | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0001817 | involved_in regulation of cytokine production | 1 | IC | Homo_sapiens(9606) | Process |
| GO:0003723 | enables RNA binding | 1 | HDA | Homo_sapiens(9606) | Function |
| GO:0003730 | enables mRNA 3-UTR binding | 2 | IBA,IDA | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005634 | is_active_in nucleus | 1 | IBA | Homo_sapiens(9606) | Component |
| GO:0005737 | is_active_in cytoplasm | 2 | IBA,IDA | Homo_sapiens(9606) | Component |
| GO:0005829 | is_active_in cytosol | 3 | IBA,IDA,TAS | Homo_sapiens(9606) | Component |
| GO:0006412 | involved_in translation | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0007399 | involved_in nervous system development | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0009653 | involved_in anatomical structure morphogenesis | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0010468 | involved_in regulation of gene expression | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0010494 | located_in cytoplasmic stress granule | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0017148 | involved_in negative regulation of translation | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0045182 | enables translation regulator activity | 1 | ISS | Homo_sapiens(9606) | Function |
| GO:0048027 | enables mRNA 5-UTR binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0051028 | involved_in mRNA transport | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0070934 | involved_in CRD-mediated mRNA stabilization | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:1990247 | enables N6-methyladenosine-containing RNA binding | 1 | IDA | Homo_sapiens(9606) | Function |