Welcome to
RBP World!
Functions and Diseases
| RBP Type | Canonical_RBPs |
| Diseases | Cancer |
| Drug | N.A. |
| Main interacting RNAs | mRNA |
| Moonlighting functions | TF |
| Localizations | Stress granucleNucleoli |
| BulkPerturb-seq | DataSet_02_130 DataSet_02_131 |
Description
| Ensembl ID | ENSG00000131914 | Gene ID | 79727 | Accession | 15986 |
| Symbol | LIN28A | Alias | CSDD1;LIN28;LIN-28;ZCCHC1;lin-28A | Full Name | lin-28 homolog A |
| Status | Confidence | Length | 18912 bases | Strand | Plus strand |
| Position | 1 : 26410817 - 26429728 | RNA binding domain | CSD , zf-CCHC | ||
| Summary | This gene encodes a LIN-28 family RNA-binding protein that acts as a posttranscriptional regulator of genes involved in developmental timing and self-renewal in embryonic stem cells. The encoded protein functions through direct interaction with target mRNAs and by disrupting the maturation of certain miRNAs involved in embryonic development. This protein prevents the terminal processing of the LET7 family of microRNAs which are major regulators of cellular growth and differentiation. Aberrant expression of this gene is associated with cancer progression in multiple tissues. [provided by RefSeq, Sep 2015] | ||||
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|---|---|---|---|
| 31883920 | LIN28B Impairs the Transition of hESC-Derived β Cells from the Juvenile to Adult State. | Xin Zhou | 2020-01-14 | Stem cell reports |
| 25457611 | Trim25 Is an RNA-Specific Activator of Lin28a/TuT4-Mediated Uridylation. | Roy Nila Choudhury | 2014-11-20 | Cell reports |
| 25368430 | MUC1-C Induces the LIN28B→LET-7→HMGA2 Axis to Regulate Self-Renewal in NSCLC. | Maroof Alam | 2015-03-01 | Molecular cancer research : MCR |
| 26481147 | A LIN28B-RAN-AURKA Signaling Network Promotes Neuroblastoma Tumorigenesis. | W Robert Schnepp | 2015-11-09 | Cancer cell |
| 29088719 | The downregulation of putative anticancer target BORIS/CTCFL in an addicted myeloid cancer cell line modulates the expression of multiple protein coding and ncRNA genes. | Evgeny Teplyakov | 2017-09-26 | Oncotarget |
| 30365085 | High LIN28A and PLK4 co‑expression is associated with poor prognosis in epithelial ovarian cancer. | Yao He | 2018-12-01 | Molecular medicine reports |
| 32601179 | LIN28B regulates transcription and potentiates MYCN-induced neuroblastoma through binding to ZNF143 at target gene promotors. | Ting Tao | 2020-07-14 | Proceedings of the National Academy of Sciences of the United States of America |
| 33665193 | Hypoxia-Induced LIN28A mRNA Promotes the Metastasis of Colon Cancer in a Protein-Coding-Independent Manner. | Mingjiao Weng | 2021-01-01 | Frontiers in cell and developmental biology |
| 27835588 | Expressions of miR-30c and let-7a are inversely correlated with HMGA2 expression in squamous cell carcinoma of the vulva. | Antonio Agostini | 2016-12-20 | Oncotarget |
| 30976203 | Prognostic value of Lin28A and Lin28B in various human malignancies: a systematic review and meta-analysis. | Jiayi Zhang | 2019-01-01 | Cancer cell international |
| 26076587 | Lhx8 regulates primordial follicle activation and postnatal folliculogenesis. | Yu Ren | 2015-06-16 | BMC biology |
| 34277782 | Lin28A promotes the proliferation and stemness of lung cancer cells via the activation of mitogen-activated protein kinase pathway dependent on microRNA let-7c. | Rui Zhang | 2021-06-01 | Annals of translational medicine |
| 23550645 | mir-17-92: a polycistronic oncomir with pleiotropic functions. | Virginie Olive | 2013-05-01 | Immunological reviews |
| 32359650 | Retinoic acid and microRNA. | Lijun Wang | 2020-01-01 | Methods in enzymology |
| 36276965 | Overexpression of LncRNA SNHG14 as a biomarker of clinicopathological and prognosis value in human cancers: A meta-analysis and bioinformatics analysis. | Bin Liu | 2022-01-01 | Frontiers in genetics |
| 22240064 | Analysis of LIN28A in early human ovary development and as a candidate gene for primary ovarian insufficiency. | Ranna El-Khairi | 2012-04-04 | Molecular and cellular endocrinology |
| 29540527 | A biosensor for MAPK-dependent Lin28 signaling. | M Laurel Oldach | 2018-05-15 | Molecular biology of the cell |
| 36230562 | Lin28 Regulates Cancer Cell Stemness for Tumour Progression. | Zhuohui Lin | 2022-09-24 | Cancers |
| 26893371 | Clinicopathological Characteristics of Patients with Gastric Cancer according to the Expression of LIN28A. | Hyuk Chan Park | 2016-09-15 | Gut and liver |
| 27494865 | Lin28A activates androgen receptor via regulation of c-myc and promotes malignancy of ER-/Her2+ breast cancer. | Honghong Shen | 2016-09-13 | Oncotarget |
| 33504840 | Identification of RNA-binding proteins that partner with Lin28a to regulate Dnmt3a expression. | Silvia Parisi | 2021-01-27 | Scientific reports |
| 26910839 | MeCP2 suppresses LIN28A expression via binding to its methylated-CpG islands in pancreatic cancer cells. | Min Xu | 2016-03-22 | Oncotarget |
| 30266988 | The interaction of Lin28A/Rho associated coiled-coil containing protein kinase2 accelerates the malignancy of ovarian cancer. | Yancheng Zhong | 2019-02-01 | Oncogene |
| 27659051 | ARiBo pull-down for riboproteomic studies based on label-free quantitative mass spectrometry. | Geneviève G Di | 2016-11-01 | RNA (New York, N.Y.) |
| 32278210 | Mechanisms of neurite repair. | Han-Hsuan Liu | 2020-08-01 | Current opinion in neurobiology |
| 22751018 | Structural basis for the activity of a cytoplasmic RNA terminal uridylyl transferase. | A Luke Yates | 2012-08-01 | Nature structural & molecular biology |
| 36509142 | RNA-binding protein LIN28A upregulates transcription factor HIF1α by posttranscriptional regulation via direct binding to UGAU motifs. | Hiroto Yamamoto | 2023-01-01 | The Journal of biological chemistry |
| 34661235 | KLF17 promotes human naïve pluripotency but is not required for its establishment. | A Rebecca Lea | 2021-11-15 | Development (Cambridge, England) |
| 35601075 | LIN28A alleviates inflammation, oxidative stress, osteogenic differentiation and mineralization in lipopolysaccharide (LPS)-treated human periodontal ligament stem cells. | Ling Guo | 2022-06-01 | Experimental and therapeutic medicine |
| 25323333 | Genome-wide profiling of mouse RNA secondary structures reveals key features of the mammalian transcriptome. | Danny Incarnato | 2014-01-01 | Genome biology |
| 27107971 | Rapid reversal of translational silencing: Emerging role of microRNA degradation pathways in neuronal plasticity. | Xiuping Fu | 2016-09-01 | Neurobiology of learning and memory |
| 36090160 | Case report: Infratentorial Embryonal Tumor with Abundant Neuropil and True Rosettes (ETANTR) in an 8-month-old Maine Coon. | Greta Foiani | 2022-01-01 | Frontiers in veterinary science |
| 24912481 | A 1536-well fluorescence polarization assay to screen for modulators of the MUSASHI family of RNA-binding proteins. | Gerard Minuesa | 2014-01-01 | Combinatorial chemistry & high throughput screening |
| 28569789 | Knockdown of miR-128a induces Lin28a expression and reverts myeloid differentiation blockage in acute myeloid leukemia. | Luciana L De | 2017-06-01 | Cell death & disease |
| 34868981 | Lin28A/CENPE Promoting the Proliferation and Chemoresistance of Acute Myeloid Leukemia. | Mingyue Shi | 2021-01-01 | Frontiers in oncology |
| 32731422 | The Emerging Role of Galectins and O-GlcNAc Homeostasis in Processes of Cellular Differentiation. | Rada Tazhitdinova | 2020-07-28 | Cells |
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| LIN28A-201 | ENST00000254231 | 3458 | 209aa | ENSP00000254231 |
| LIN28A-202 | ENST00000326279 | 3975 | 209aa | ENSP00000363314 |
Phenotypes
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| 48432 | Child Development Disorders, Pervasive | 2.1091700E-005 | - |
| 87854 | Body Height | 3E-8 | 20881960 |
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
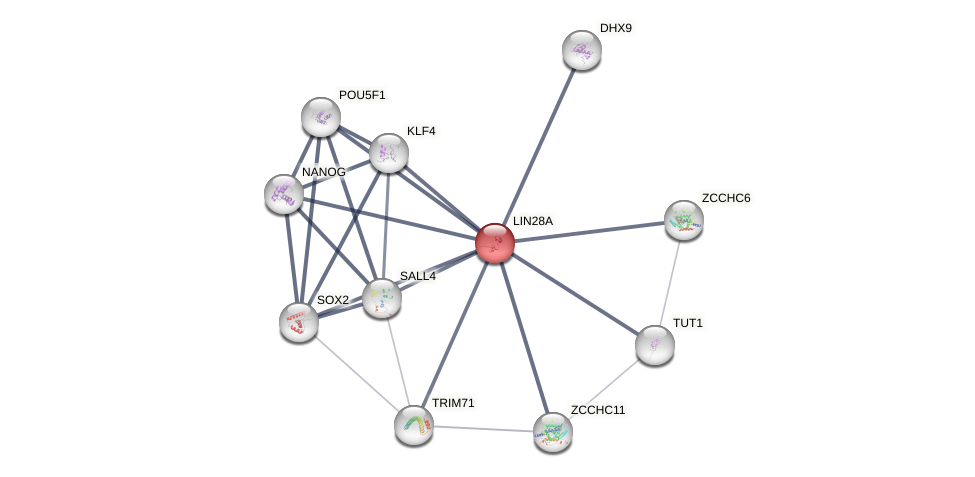
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000131914 | homo_sapiens | ENSP00000007699 | 22.8022 | 16.7582 | homo_sapiens | ENSP00000363314 | 39.7129 | 29.1866 |
| ENSG00000131914 | homo_sapiens | ENSP00000361626 | 32.716 | 23.1481 | homo_sapiens | ENSP00000363314 | 50.7177 | 35.8852 |
| ENSG00000131914 | homo_sapiens | ENSP00000228251 | 23.3871 | 16.129 | homo_sapiens | ENSP00000363314 | 41.6268 | 28.7081 |
| ENSG00000131914 | homo_sapiens | ENSP00000344401 | 60.4 | 54.4 | homo_sapiens | ENSP00000363314 | 72.2488 | 65.0718 |
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 99.0431 | 98.0861 | nomascus_leucogenys | ENSNLEP00000028611 | 99.0431 | 98.0861 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 100 | 99.0431 | pan_paniscus | ENSPPAP00000029767 | 100 | 99.0431 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 99.5215 | 99.0431 | pongo_abelii | ENSPPYP00000001946 | 99.5215 | 99.0431 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 100 | 99.5215 | pan_troglodytes | ENSPTRP00000088583 | 100 | 99.5215 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 100 | 99.5215 | gorilla_gorilla | ENSGGOP00000044505 | 100 | 99.5215 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 87.0813 | 82.7751 | ornithorhynchus_anatinus | ENSOANP00000037351 | 87.5 | 83.1731 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 93.7799 | 90.4306 | sarcophilus_harrisii | ENSSHAP00000032502 | 95.6098 | 92.1951 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 93.7799 | 90.4306 | notamacropus_eugenii | ENSMEUP00000007981 | 95.6098 | 92.1951 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 65.5502 | 64.1148 | choloepus_hoffmanni | ENSCHOP00000001832 | 66.8293 | 65.3659 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 92.823 | 90.4306 | dasypus_novemcinctus | ENSDNOP00000008289 | 94.6341 | 92.1951 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 76.555 | 68.8995 | dasypus_novemcinctus | ENSDNOP00000018122 | 82.4742 | 74.2268 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 90.4306 | 89.4737 | erinaceus_europaeus | ENSEEUP00000011030 | 95.4545 | 94.4444 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 99.5215 | 99.0431 | callithrix_jacchus | ENSCJAP00000017956 | 99.5215 | 99.0431 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 99.5215 | 99.0431 | cercocebus_atys | ENSCATP00000006763 | 99.5215 | 99.0431 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 99.5215 | 99.0431 | macaca_fascicularis | ENSMFAP00000062993 | 99.5215 | 99.0431 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 99.0431 | 98.5646 | macaca_nemestrina | ENSMNEP00000024673 | 99.0431 | 98.5646 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 99.0431 | 98.5646 | papio_anubis | ENSPANP00000001729 | 99.0431 | 98.5646 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 99.5215 | 99.0431 | mandrillus_leucophaeus | ENSMLEP00000002829 | 99.5215 | 99.0431 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 89.4737 | 87.0813 | tursiops_truncatus | ENSTTRP00000005361 | 96.3918 | 93.8144 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 71.7703 | 65.0718 | vulpes_vulpes | ENSVVUP00000017807 | 60.4839 | 54.8387 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 98.5646 | 96.6507 | mus_spretus | MGP_SPRETEiJ_P0067542 | 98.5646 | 96.6507 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 94.7368 | 91.3876 | equus_asinus | ENSEASP00005041942 | 96.5854 | 93.1707 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 79.4258 | 74.6412 | capra_hircus | ENSCHIP00000032642 | 85.567 | 80.4124 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 94.7368 | 92.823 | loxodonta_africana | ENSLAFP00000014696 | 96.5854 | 94.6341 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 96.1722 | 94.7368 | panthera_leo | ENSPLOP00000014570 | 97.5728 | 96.1165 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 96.1722 | 94.7368 | ailuropoda_melanoleuca | ENSAMEP00000011131 | 97.5728 | 96.1165 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 85.6459 | 79.9043 | bos_indicus_hybrid | ENSBIXP00005000822 | 83.6449 | 78.0374 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 86.1244 | 83.7321 | oryctolagus_cuniculus | ENSOCUP00000041421 | 95.2381 | 92.5926 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 98.0861 | 96.1722 | oryctolagus_cuniculus | ENSOCUP00000023367 | 98.0861 | 96.1722 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 98.0861 | 96.1722 | oryctolagus_cuniculus | ENSOCUP00000000262 | 98.0861 | 96.1722 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 94.7368 | 91.3876 | equus_caballus | ENSECAP00000017733 | 96.5854 | 93.1707 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 46.4115 | 43.0622 | canis_lupus_familiaris | ENSCAFP00845034036 | 69.7842 | 64.7482 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 96.1722 | 94.7368 | panthera_pardus | ENSPPRP00000017123 | 97.5728 | 96.1165 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 73.6842 | 68.8995 | marmota_marmota_marmota | ENSMMMP00000015780 | 60.3922 | 56.4706 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 93.3014 | 90.4306 | balaenoptera_musculus | ENSBMSP00010018194 | 94.6602 | 91.7476 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 95.2153 | 92.823 | balaenoptera_musculus | ENSBMSP00010008897 | 97.0732 | 94.6341 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 97.6077 | 94.2584 | cavia_porcellus | ENSCPOP00000019063 | 97.6077 | 94.2584 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 67.9426 | 65.0718 | cavia_porcellus | ENSCPOP00000015653 | 96.5986 | 92.517 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 92.823 | 90.4306 | felis_catus | ENSFCAP00000054902 | 83.6207 | 81.4655 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 95.6938 | 94.2584 | ursus_americanus | ENSUAMP00000008207 | 97.0874 | 95.6311 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 93.7799 | 90.9091 | monodelphis_domestica | ENSMODP00000017794 | 95.6098 | 92.6829 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 92.3445 | 88.9952 | bison_bison_bison | ENSBBBP00000011846 | 96.0199 | 92.5373 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 95.2153 | 92.3445 | bison_bison_bison | ENSBBBP00000020431 | 97.0732 | 94.1463 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 86.1244 | 83.7321 | monodon_monoceros | ENSMMNP00015012951 | 93.2643 | 90.6736 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 78.4689 | 74.6412 | monodon_monoceros | ENSMMNP00015007941 | 82.4121 | 78.392 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 95.2153 | 92.823 | monodon_monoceros | ENSMMNP00015008144 | 97.0732 | 94.6341 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 96.1722 | 93.3014 | sus_scrofa | ENSSSCP00000003860 | 98.0488 | 95.1219 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 88.5167 | 87.0813 | microcebus_murinus | ENSMICP00000022563 | 96.3542 | 94.7917 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 95.2153 | 91.3876 | octodon_degus | ENSODEP00000005682 | 96.1353 | 92.2705 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 94.7368 | 92.3445 | octodon_degus | ENSODEP00000025924 | 94.7368 | 92.3445 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 77.512 | 71.7703 | octodon_degus | ENSODEP00000021854 | 91.0112 | 84.2697 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 87.5598 | 82.2967 | jaculus_jaculus | ENSJJAP00000017177 | 94.8186 | 89.1192 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 82.2967 | 78.4689 | jaculus_jaculus | ENSJJAP00000000827 | 91.4894 | 87.234 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 86.1244 | 82.2967 | ovis_aries_rambouillet | ENSOARP00020027449 | 81.448 | 77.8281 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 95.6938 | 94.2584 | ursus_maritimus | ENSUMAP00000017132 | 97.0874 | 95.6311 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 98.0861 | 95.6938 | ochotona_princeps | ENSOPRP00000001452 | 98.0861 | 95.6938 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 78.4689 | 74.6412 | delphinapterus_leucas | ENSDLEP00000026578 | 82.4121 | 78.392 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 95.2153 | 92.823 | delphinapterus_leucas | ENSDLEP00000012153 | 97.0732 | 94.6341 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 89.4737 | 87.0813 | physeter_catodon | ENSPCTP00005012251 | 92.1182 | 89.6552 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 91.866 | 88.5167 | physeter_catodon | ENSPCTP00005005211 | 93.6585 | 90.2439 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 95.2153 | 92.823 | physeter_catodon | ENSPCTP00005007205 | 97.0732 | 94.6341 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 98.0861 | 96.1722 | mus_caroli | MGP_CAROLIEiJ_P0065007 | 98.0861 | 96.1722 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 98.5646 | 96.6507 | mus_musculus | ENSMUSP00000050488 | 98.5646 | 96.6507 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 70.3349 | 66.0287 | dipodomys_ordii | ENSDORP00000018507 | 81.6667 | 76.6667 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 95.6938 | 94.2584 | mustela_putorius_furo | ENSMPUP00000015615 | 97.0874 | 95.6311 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 99.0431 | 98.5646 | aotus_nancymaae | ENSANAP00000005892 | 99.0431 | 98.5646 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 94.2584 | 93.3014 | saimiri_boliviensis_boliviensis | ENSSBOP00000034205 | 97.5247 | 96.5347 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 57.8947 | 56.4593 | vicugna_pacos | ENSVPAP00000007512 | 99.1803 | 96.7213 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 99.5215 | 99.0431 | chlorocebus_sabaeus | ENSCSAP00000017034 | 99.0476 | 98.5714 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 97.6077 | 95.2153 | mesocricetus_auratus | ENSMAUP00000020996 | 97.6077 | 95.2153 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 95.2153 | 92.823 | phocoena_sinus | ENSPSNP00000006785 | 97.0732 | 94.6341 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 88.5167 | 86.1244 | phocoena_sinus | ENSPSNP00000019761 | 91.5842 | 89.1089 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 95.2153 | 92.3445 | phocoena_sinus | ENSPSNP00000003489 | 97.0732 | 94.1463 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 95.6938 | 93.3014 | camelus_dromedarius | ENSCDRP00005008412 | 97.561 | 95.1219 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 86.1244 | 84.689 | panthera_tigris_altaica | ENSPTIP00000016125 | 85.7143 | 84.2857 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 99.0431 | 98.0861 | rattus_norvegicus | ENSRNOP00000085321 | 99.0431 | 98.0861 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 99.0431 | 98.0861 | prolemur_simus | ENSPSMP00000016464 | 99.0431 | 98.0861 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 97.6077 | 95.2153 | microtus_ochrogaster | ENSMOCP00000007916 | 97.6077 | 95.2153 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 50.2392 | 48.8038 | sorex_araneus | ENSSARP00000011857 | 79.5455 | 77.2727 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 97.1292 | 94.2584 | peromyscus_maniculatus_bairdii | ENSPEMP00000011927 | 97.1292 | 94.2584 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 91.3876 | 88.0383 | bos_mutus | ENSBMUP00000005823 | 95.0249 | 91.5423 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 95.2153 | 92.823 | bos_mutus | ENSBMUP00000021827 | 97.0732 | 94.6341 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 98.5646 | 96.6507 | mus_spicilegus | ENSMSIP00000001515 | 98.5646 | 96.6507 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 98.0861 | 95.2153 | cricetulus_griseus_chok1gshd | ENSCGRP00001017024 | 98.0861 | 95.2153 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 92.3445 | 90.4306 | pteropus_vampyrus | ENSPVAP00000016289 | 97.4747 | 95.4545 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 51.6746 | 45.933 | myotis_lucifugus | ENSMLUP00000020726 | 75 | 66.6667 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 94.2584 | 90.9091 | vombatus_ursinus | ENSVURP00010008796 | 96.0976 | 92.6829 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 96.1722 | 94.7368 | rhinolophus_ferrumequinum | ENSRFEP00010005541 | 98.0488 | 96.5854 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 95.6938 | 93.7799 | neovison_vison | ENSNVIP00000030720 | 97.0874 | 95.1456 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 83.7321 | 79.4258 | bos_taurus | ENSBTAP00000065444 | 88.3838 | 83.8384 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 93.3014 | 88.9952 | heterocephalus_glaber_female | ENSHGLP00000018300 | 92.4171 | 88.1517 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 80.8612 | 76.0766 | heterocephalus_glaber_female | ENSHGLP00000001330 | 65.251 | 61.39 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 97.6077 | 95.2153 | heterocephalus_glaber_female | ENSHGLP00000045526 | 86.076 | 83.9662 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 98.5646 | 98.0861 | rhinopithecus_bieti | ENSRBIP00000015823 | 98.5646 | 98.0861 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 96.1722 | 94.7368 | canis_lupus_dingo | ENSCAFP00020027286 | 97.5728 | 96.1165 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 97.6077 | 95.6938 | sciurus_vulgaris | ENSSVLP00005003029 | 97.6077 | 95.6938 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 93.7799 | 90.9091 | phascolarctos_cinereus | ENSPCIP00000016449 | 95.6098 | 92.6829 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 90.9091 | 89.4737 | procavia_capensis | ENSPCAP00000012749 | 93.1373 | 91.6667 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 98.0861 | 96.1722 | nannospalax_galili | ENSNGAP00000018831 | 98.0861 | 96.1722 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 64.5933 | 57.4163 | bos_grunniens | ENSBGRP00000035767 | 56.0166 | 49.7925 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 97.6077 | 94.7368 | chinchilla_lanigera | ENSCLAP00000019690 | 97.6077 | 94.7368 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 95.6938 | 92.823 | cervus_hanglu_yarkandensis | ENSCHYP00000007494 | 97.561 | 94.6341 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 93.3014 | 90.9091 | cervus_hanglu_yarkandensis | ENSCHYP00000020930 | 97.0149 | 94.5274 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 95.6938 | 93.3014 | cervus_hanglu_yarkandensis | ENSCHYP00000002490 | 97.561 | 95.1219 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 95.6938 | 92.823 | catagonus_wagneri | ENSCWAP00000010114 | 97.561 | 94.6341 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 81.8182 | 80.3828 | ictidomys_tridecemlineatus | ENSSTOP00000031278 | 91.9355 | 90.3226 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 97.1292 | 95.2153 | ictidomys_tridecemlineatus | ENSSTOP00000003875 | 96.6667 | 94.7619 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 97.6077 | 97.1292 | otolemur_garnettii | ENSOGAP00000003536 | 97.6077 | 97.1292 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 99.0431 | 98.5646 | cebus_imitator | ENSCCAP00000002079 | 99.0431 | 98.5646 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 95.6938 | 93.3014 | moschus_moschiferus | ENSMMSP00000006727 | 97.561 | 95.1219 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 98.5646 | 98.0861 | rhinopithecus_roxellana | ENSRROP00000003503 | 98.5646 | 98.0861 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 89.9522 | 87.5598 | mus_pahari | MGP_PahariEiJ_P0080096 | 98.4293 | 95.8115 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 80.3828 | 77.512 | propithecus_coquereli | ENSPCOP00000009572 | 95.4545 | 92.0455 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 98.0861 | 97.6077 | propithecus_coquereli | ENSPCOP00000027975 | 98.0861 | 97.6077 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 35.8852 | 24.4019 | caenorhabditis_elegans | Y39A1C.3.1 | 25.5102 | 17.3469 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 32.5359 | 20.0957 | caenorhabditis_elegans | F46F11.2.1 | 25.4682 | 15.7303 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 33.4928 | 21.5311 | caenorhabditis_elegans | M01E11.5.1 | 26.4151 | 16.9811 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 47.8469 | 36.3636 | drosophila_melanogaster | FBpp0076798 | 51.2821 | 38.9744 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 51.1962 | 39.2345 | ciona_intestinalis | ENSCINP00000032715 | 55.1546 | 42.268 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 55.0239 | 41.6268 | eptatretus_burgeri | ENSEBUP00000013159 | 57.2139 | 43.2836 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 56.9378 | 49.2823 | callorhinchus_milii | ENSCMIP00000044739 | 66.1111 | 57.2222 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 83.2536 | 75.1196 | latimeria_chalumnae | ENSLACP00000003069 | 84.8781 | 76.5854 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 84.689 | 76.555 | lepisosteus_oculatus | ENSLOCP00000003958 | 87.1921 | 78.8177 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 78.4689 | 71.2919 | clupea_harengus | ENSCHAP00000004440 | 81.1881 | 73.7624 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 73.2057 | 63.1579 | clupea_harengus | ENSCHAP00000035702 | 74.2718 | 64.0777 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 75.5981 | 65.5502 | danio_rerio | ENSDARP00000089741 | 78.2178 | 67.8218 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 76.555 | 66.9856 | danio_rerio | ENSDARP00000138197 | 82.9016 | 72.5389 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 76.555 | 68.4211 | carassius_auratus | ENSCARP00000039269 | 82.9016 | 74.0933 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 76.0766 | 68.4211 | carassius_auratus | ENSCARP00000041808 | 77.1845 | 69.4175 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 72.7273 | 61.244 | carassius_auratus | ENSCARP00000052132 | 77.1574 | 64.9746 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 77.0335 | 63.6364 | carassius_auratus | ENSCARP00000098299 | 76.6667 | 63.3333 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 80.8612 | 72.2488 | astyanax_mexicanus | ENSAMXP00000040941 | 82.0388 | 73.301 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 74.1627 | 64.1148 | astyanax_mexicanus | ENSAMXP00000030871 | 73.4597 | 63.5071 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 73.2057 | 63.6364 | ictalurus_punctatus | ENSIPUP00000017059 | 79.2746 | 68.9119 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 78.9474 | 71.2919 | ictalurus_punctatus | ENSIPUP00000022070 | 81.2808 | 73.399 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 77.9904 | 70.8134 | esox_lucius | ENSELUP00000038209 | 81.5 | 74 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 74.1627 | 64.5933 | esox_lucius | ENSELUP00000032280 | 71.1009 | 61.9266 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 74.6412 | 64.5933 | electrophorus_electricus | ENSEEEP00000028355 | 76.8473 | 66.5025 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 77.0335 | 67.4641 | electrophorus_electricus | ENSEEEP00000051160 | 77.0335 | 67.4641 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 74.6412 | 66.5072 | oncorhynchus_kisutch | ENSOKIP00005048145 | 77.2277 | 68.8119 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 74.1627 | 67.4641 | oncorhynchus_kisutch | ENSOKIP00005079005 | 74.8792 | 68.1159 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 79.4258 | 71.7703 | oncorhynchus_kisutch | ENSOKIP00005046246 | 82.1782 | 74.2574 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 80.3828 | 72.7273 | oncorhynchus_kisutch | ENSOKIP00005018537 | 83.1683 | 75.2475 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 72.2488 | 64.5933 | oncorhynchus_mykiss | ENSOMYP00000117184 | 77.4359 | 69.2308 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 74.1627 | 65.0718 | oncorhynchus_mykiss | ENSOMYP00000069020 | 75.9804 | 66.6667 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 79.9043 | 72.2488 | oncorhynchus_mykiss | ENSOMYP00000025281 | 82.6733 | 74.7525 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 79.9043 | 72.2488 | oncorhynchus_mykiss | ENSOMYP00000140576 | 82.6733 | 74.7525 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 74.1627 | 65.0718 | salmo_salar | ENSSSAP00000144406 | 75.9804 | 66.6667 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 80.3828 | 72.7273 | salmo_salar | ENSSSAP00000151601 | 83.1683 | 75.2475 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 75.1196 | 67.9426 | salmo_salar | ENSSSAP00000132646 | 77.7228 | 70.297 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 80.3828 | 72.7273 | salmo_salar | ENSSSAP00000153321 | 83.1683 | 75.2475 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 79.9043 | 72.2488 | salmo_trutta | ENSSTUP00000103841 | 82.6733 | 74.7525 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 75.5981 | 68.4211 | salmo_trutta | ENSSTUP00000109147 | 75.9615 | 68.75 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 80.8612 | 73.2057 | salmo_trutta | ENSSTUP00000015050 | 83.6634 | 75.7426 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 74.6412 | 66.5072 | salmo_trutta | ENSSTUP00000091977 | 77.2277 | 68.8119 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 78.4689 | 71.2919 | gadus_morhua | ENSGMOP00000024802 | 81.1881 | 73.7624 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 67.4641 | 58.3732 | gadus_morhua | ENSGMOP00000047301 | 71.5736 | 61.9289 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 78.9474 | 72.2488 | fundulus_heteroclitus | ENSFHEP00000000321 | 81.6832 | 74.7525 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 78.4689 | 71.7703 | poecilia_reticulata | ENSPREP00000007670 | 81.1881 | 74.2574 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 78.9474 | 72.2488 | xiphophorus_maculatus | ENSXMAP00000014615 | 81.6832 | 74.7525 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 78.4689 | 71.2919 | oryzias_latipes | ENSORLP00000042231 | 82.4121 | 74.8744 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 79.4258 | 72.2488 | cyclopterus_lumpus | ENSCLMP00005013179 | 82.5871 | 75.1244 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 70.3349 | 61.244 | cyclopterus_lumpus | ENSCLMP00005017342 | 71.7073 | 62.439 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 78.4689 | 70.8134 | oreochromis_niloticus | ENSONIP00000020405 | 81.1881 | 73.2673 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 77.9904 | 70.8134 | haplochromis_burtoni | ENSHBUP00000007778 | 80.6931 | 73.2673 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 77.9904 | 70.8134 | astatotilapia_calliptera | ENSACLP00000041250 | 80.6931 | 73.2673 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 68.4211 | 58.8517 | sparus_aurata | ENSSAUP00010032583 | 74.0933 | 63.7306 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 79.9043 | 71.2919 | sparus_aurata | ENSSAUP00010041805 | 82.6733 | 73.7624 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 71.2919 | 61.7225 | lates_calcarifer | ENSLCAP00010034396 | 67.7273 | 58.6364 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 79.9043 | 73.6842 | lates_calcarifer | ENSLCAP00010024315 | 82.6733 | 76.2376 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 72.7273 | 63.1579 | lates_calcarifer | ENSLCAP00010006659 | 70.0461 | 60.8295 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 72.2488 | 63.1579 | xenopus_tropicalis | ENSXETP00000112624 | 77.4359 | 67.6923 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 89.9522 | 81.8182 | chrysemys_picta_bellii | ENSCPBP00000035963 | 92.1569 | 83.8235 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 88.9952 | 82.2967 | sphenodon_punctatus | ENSSPUP00000013278 | 90.7317 | 83.9024 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 67.4641 | 62.201 | crocodylus_porosus | ENSCPRP00005005851 | 71.5736 | 65.9898 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 85.6459 | 78.4689 | laticauda_laticaudata | ENSLLTP00000007727 | 89.5 | 82 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 88.0383 | 81.8182 | notechis_scutatus | ENSNSUP00000018982 | 89.3204 | 83.0097 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 83.7321 | 77.512 | pseudonaja_textilis | ENSPTXP00000007581 | 86.6337 | 80.198 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 67.9426 | 63.1579 | anas_platyrhynchos_platyrhynchos | ENSAPLP00000017925 | 71 | 66 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 87.5598 | 81.8182 | gallus_gallus | ENSGALP00010023368 | 79.2208 | 74.026 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 86.1244 | 80.3828 | meleagris_gallopavo | ENSMGAP00000009761 | 89.1089 | 83.1683 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 71.2919 | 64.1148 | serinus_canaria | ENSSCAP00000009341 | 60.0806 | 54.0323 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 87.0813 | 80.8612 | parus_major | ENSPMJP00000024470 | 90.099 | 83.6634 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 59.3301 | 55.0239 | anolis_carolinensis | ENSACAP00000017489 | 93.2331 | 86.4662 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 77.9904 | 70.3349 | neolamprologus_brichardi | ENSNBRP00000012097 | 80.6931 | 72.7723 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 83.7321 | 77.0335 | naja_naja | ENSNNAP00000012481 | 87.9397 | 80.9045 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 88.0383 | 80.8612 | podarcis_muralis | ENSPMRP00000014508 | 88.8889 | 81.6425 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 64.1148 | 56.4593 | geospiza_fortis | ENSGFOP00000010588 | 62.9108 | 55.3991 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 86.6029 | 80.3828 | taeniopygia_guttata | ENSTGUP00000019295 | 89.604 | 83.1683 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 77.9904 | 70.3349 | erpetoichthys_calabaricus | ENSECRP00000023527 | 82.7411 | 74.6193 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 79.9043 | 72.7273 | kryptolebias_marmoratus | ENSKMAP00000018031 | 82.6733 | 75.2475 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 88.9952 | 82.2967 | salvator_merianae | ENSSMRP00000002611 | 89.8551 | 83.0918 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 78.4689 | 71.2919 | oryzias_javanicus | ENSOJAP00000006276 | 82.4121 | 74.8744 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 78.4689 | 70.8134 | amphilophus_citrinellus | ENSACIP00000022253 | 78.8462 | 71.1538 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 80.3828 | 73.6842 | dicentrarchus_labrax | ENSDLAP00005073587 | 83.1683 | 76.2376 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 70.3349 | 60.7655 | dicentrarchus_labrax | ENSDLAP00005075245 | 78.1915 | 67.5532 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 79.4258 | 72.2488 | amphiprion_percula | ENSAPEP00000008635 | 79.8077 | 72.5962 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 67.4641 | 59.3301 | amphiprion_percula | ENSAPEP00000010429 | 67.7885 | 59.6154 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 60.7655 | 56.4593 | pelodiscus_sinensis | ENSPSIP00000020326 | 84.106 | 78.1457 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 89.9522 | 81.8182 | gopherus_evgoodei | ENSGEVP00005030408 | 92.1569 | 83.8235 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 89.4737 | 81.3397 | chelonoidis_abingdonii | ENSCABP00000014125 | 91.6667 | 83.3333 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 70.8134 | 58.8517 | seriola_dumerili | ENSSDUP00000015967 | 83.1461 | 69.1011 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 76.555 | 70.3349 | seriola_dumerili | ENSSDUP00000013389 | 82.0513 | 75.3846 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 79.9043 | 72.2488 | cottoperca_gobio | ENSCGOP00000025662 | 82.266 | 74.3842 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 85.1675 | 78.4689 | anser_brachyrhynchus | ENSABRP00000001359 | 87.2549 | 80.3922 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 64.5933 | 55.5024 | gasterosteus_aculeatus | ENSGACP00000010372 | 80.3571 | 69.0476 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 75.1196 | 68.4211 | gasterosteus_aculeatus | ENSGACP00000018348 | 77.3399 | 70.4434 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 78.4689 | 71.7703 | cynoglossus_semilaevis | ENSCSEP00000029793 | 81.1881 | 74.2574 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 78.9474 | 71.7703 | poecilia_formosa | ENSPFOP00000014075 | 81.6832 | 74.2574 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 72.2488 | 63.6364 | myripristis_murdjan | ENSMMDP00005016136 | 81.6216 | 71.8919 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 80.8612 | 73.2057 | myripristis_murdjan | ENSMMDP00005027914 | 83.6634 | 75.7426 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 79.4258 | 72.2488 | sander_lucioperca | ENSSLUP00000049422 | 82.1782 | 74.7525 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 71.7703 | 62.6794 | sander_lucioperca | ENSSLUP00000005378 | 72.8155 | 63.5922 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 86.6029 | 80.8612 | strigops_habroptila | ENSSHBP00005019766 | 89.604 | 83.6634 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 79.4258 | 72.2488 | amphiprion_ocellaris | ENSAOCP00000016552 | 79.8077 | 72.5962 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 68.4211 | 60.2871 | amphiprion_ocellaris | ENSAOCP00000026015 | 68.4211 | 60.2871 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 89.9522 | 82.2967 | terrapene_carolina_triunguis | ENSTMTP00000016596 | 92.1569 | 84.3137 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 77.512 | 69.378 | hucho_hucho | ENSHHUP00000025445 | 73.6364 | 65.9091 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 73.6842 | 65.0718 | hucho_hucho | ENSHHUP00000085533 | 78.9744 | 69.7436 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 76.0766 | 67.9426 | hucho_hucho | ENSHHUP00000066081 | 77.561 | 69.2683 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 69.8565 | 61.7225 | hucho_hucho | ENSHHUP00000072376 | 73.3668 | 64.8241 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 79.4258 | 73.2057 | betta_splendens | ENSBSLP00000048341 | 82.5871 | 76.1194 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 64.1148 | 53.5885 | betta_splendens | ENSBSLP00000029298 | 69.4301 | 58.0311 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 76.0766 | 65.0718 | pygocentrus_nattereri | ENSPNAP00000013141 | 79.1045 | 67.6617 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 79.4258 | 71.7703 | pygocentrus_nattereri | ENSPNAP00000029542 | 80.9756 | 73.1707 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 71.2919 | 62.201 | anabas_testudineus | ENSATEP00000012704 | 73.7624 | 64.3564 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 80.3828 | 73.6842 | anabas_testudineus | ENSATEP00000014616 | 83.1683 | 76.2376 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 47.3684 | 32.5359 | ciona_savignyi | ENSCSAVP00000011162 | 56.5714 | 38.8571 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 76.0766 | 69.8565 | seriola_lalandi_dorsalis | ENSSLDP00000003983 | 81.5385 | 74.8718 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 73.2057 | 61.7225 | seriola_lalandi_dorsalis | ENSSLDP00000026493 | 75 | 63.2353 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 68.4211 | 58.8517 | labrus_bergylta | ENSLBEP00000024327 | 70.7921 | 60.8911 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 68.4211 | 58.8517 | labrus_bergylta | ENSLBEP00000001133 | 70.7921 | 60.8911 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 77.512 | 67.4641 | labrus_bergylta | ENSLBEP00000036852 | 77.1429 | 67.1429 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 77.9904 | 70.8134 | pundamilia_nyererei | ENSPNYP00000018595 | 78.3654 | 71.1538 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 80.8612 | 73.6842 | mastacembelus_armatus | ENSMAMP00000010781 | 83.6634 | 76.2376 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 72.7273 | 63.6364 | mastacembelus_armatus | ENSMAMP00000002925 | 75.2475 | 65.8416 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 79.9043 | 73.2057 | stegastes_partitus | ENSSPAP00000016682 | 82.6733 | 75.7426 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 67.4641 | 58.8517 | stegastes_partitus | ENSSPAP00000024845 | 73.057 | 63.7306 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 78.9474 | 71.2919 | oncorhynchus_tshawytscha | ENSOTSP00005015205 | 81.6832 | 73.7624 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 78.9474 | 71.2919 | oncorhynchus_tshawytscha | ENSOTSP00005069620 | 81.6832 | 73.7624 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 80.3828 | 72.7273 | oncorhynchus_tshawytscha | ENSOTSP00005053819 | 83.1683 | 75.2475 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 75.5981 | 67.9426 | oncorhynchus_tshawytscha | ENSOTSP00005093596 | 75.9615 | 68.2692 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 74.6412 | 66.5072 | oncorhynchus_tshawytscha | ENSOTSP00005075764 | 77.2277 | 68.8119 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 78.9474 | 71.2919 | oncorhynchus_tshawytscha | ENSOTSP00005000080 | 81.6832 | 73.7624 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 80.3828 | 72.7273 | oncorhynchus_tshawytscha | ENSOTSP00005013370 | 83.1683 | 75.2475 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 76.555 | 67.9426 | acanthochromis_polyacanthus | ENSAPOP00000020652 | 74.7664 | 66.3551 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 65.0718 | 57.4163 | acanthochromis_polyacanthus | ENSAPOP00000011727 | 71.5789 | 63.1579 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 86.1244 | 80.3828 | aquila_chrysaetos_chrysaetos | ENSACCP00020013752 | 90 | 84 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 79.4258 | 72.7273 | nothobranchius_furzeri | ENSNFUP00015001975 | 81.3726 | 74.5098 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 78.4689 | 70.8134 | cyprinodon_variegatus | ENSCVAP00000029493 | 81.1881 | 73.2673 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 72.2488 | 65.5502 | denticeps_clupeoides | ENSDCDP00000025623 | 81.1828 | 73.6559 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 77.512 | 69.378 | denticeps_clupeoides | ENSDCDP00000022951 | 64.8 | 58 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 80.3828 | 73.6842 | larimichthys_crocea | ENSLCRP00005030312 | 83.1683 | 76.2376 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 72.2488 | 63.1579 | larimichthys_crocea | ENSLCRP00005018988 | 69.2661 | 60.5505 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 77.512 | 68.4211 | takifugu_rubripes | ENSTRUP00000067644 | 70.7424 | 62.4454 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 66.5072 | 56.4593 | takifugu_rubripes | ENSTRUP00000084237 | 67.8049 | 57.561 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 83.2536 | 75.5981 | ficedula_albicollis | ENSFALP00000018195 | 86.5672 | 78.607 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 78.9474 | 70.8134 | hippocampus_comes | ENSHCOP00000005033 | 78.9474 | 70.8134 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 77.9904 | 66.5072 | cyprinus_carpio_carpio | ENSCCRP00000104483 | 78.3654 | 66.8269 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 77.0335 | 65.0718 | cyprinus_carpio_carpio | ENSCCRP00000142605 | 77.4038 | 65.3846 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 75.5981 | 67.4641 | cyprinus_carpio_carpio | ENSCCRP00000063300 | 67.8112 | 60.515 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 77.0335 | 68.4211 | cyprinus_carpio_carpio | ENSCCRP00000033721 | 82.1429 | 72.9592 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 86.6029 | 80.3828 | coturnix_japonica | ENSCJPP00005019036 | 89.604 | 83.1683 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 78.9474 | 71.7703 | poecilia_latipinna | ENSPLAP00000030081 | 81.6832 | 74.2574 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 77.0335 | 66.0287 | sinocyclocheilus_grahami | ENSSGRP00000107615 | 77.4038 | 66.3462 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 54.5455 | 47.3684 | sinocyclocheilus_grahami | ENSSGRP00000018162 | 69.9387 | 60.7362 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 78.9474 | 69.378 | sinocyclocheilus_grahami | ENSSGRP00000043054 | 77.4648 | 68.0751 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 77.9904 | 70.8134 | maylandia_zebra | ENSMZEP00005009120 | 80.6931 | 73.2673 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 63.1579 | 52.6316 | tetraodon_nigroviridis | ENSTNIP00000012690 | 80.9816 | 67.4847 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 25.8373 | 22.9665 | tetraodon_nigroviridis | ENSTNIP00000008017 | 73.9726 | 65.7534 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 78.9474 | 71.2919 | paramormyrops_kingsleyae | ENSPKIP00000019312 | 84.1837 | 76.0204 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 77.9904 | 70.8134 | scleropages_formosus | ENSSFOP00015044758 | 80.6931 | 73.2673 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 68.4211 | 61.244 | leptobrachium_leishanense | ENSLLEP00000010651 | 72.5888 | 64.9746 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 68.4211 | 58.8517 | scophthalmus_maximus | ENSSMAP00000045481 | 73.3333 | 63.0769 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 79.9043 | 72.2488 | scophthalmus_maximus | ENSSMAP00000008497 | 79.5238 | 71.9048 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 75.5981 | 67.9426 | oryzias_melastigma | ENSOMEP00000016853 | 73.8318 | 66.3551 |
| ENSG00000131914 | homo_sapiens | ENSP00000363314 | 46.4115 | 41.6268 | oryzias_melastigma | ENSOMEP00000034656 | 70.2899 | 63.0435 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0000932 | located_in P-body | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0002151 | enables G-quadruplex RNA binding | 1 | ISS | Homo_sapiens(9606) | Function |
| GO:0003723 | enables RNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0003729 | enables mRNA binding | 1 | IBA | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005634 | is_active_in nucleus | 2 | IBA,IDA | Homo_sapiens(9606) | Component |
| GO:0005730 | located_in nucleolus | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005737 | is_active_in cytoplasm | 2 | IBA,IDA | Homo_sapiens(9606) | Component |
| GO:0005791 | located_in rough endoplasmic reticulum | 1 | ISS | Homo_sapiens(9606) | Component |
| GO:0005829 | located_in cytosol | 2 | IDA,TAS | Homo_sapiens(9606) | Component |
| GO:0005844 | part_of polysome | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0007281 | involved_in germ cell development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0008270 | enables zinc ion binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0010494 | located_in cytoplasmic stress granule | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0010587 | involved_in miRNA catabolic process | 2 | IMP,ISS | Homo_sapiens(9606) | Process |
| GO:0017148 | involved_in negative regulation of translation | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0019827 | involved_in stem cell population maintenance | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0031054 | involved_in pre-miRNA processing | 2 | IBA,IMP | Homo_sapiens(9606) | Process |
| GO:0031123 | involved_in RNA 3-end processing | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0031369 | enables translation initiation factor binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0032008 | involved_in positive regulation of TOR signaling | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0035198 | enables miRNA binding | 1 | ISS | Homo_sapiens(9606) | Function |
| GO:0045666 | involved_in positive regulation of neuron differentiation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0045686 | involved_in negative regulation of glial cell differentiation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0048863 | involved_in stem cell differentiation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0051897 | involved_in positive regulation of protein kinase B signaling | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0070883 | enables pre-miRNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0071076 | involved_in RNA 3 uridylation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0071333 | involved_in cellular response to glucose stimulus | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0140517 | enables protein-RNA adaptor activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:1901724 | involved_in positive regulation of cell proliferation involved in kidney development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:1905538 | enables polysome binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:1990825 | enables sequence-specific mRNA binding | 1 | ISS | Homo_sapiens(9606) | Function |
| GO:1990904 | part_of ribonucleoprotein complex | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:2000632 | involved_in negative regulation of pre-miRNA processing | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:2000767 | involved_in positive regulation of cytoplasmic translation | 1 | IMP | Homo_sapiens(9606) | Process |