Welcome to
RBP World!
Functions and Diseases
| RBP Type | Canonical_RBPs |
| Diseases | CancerCHIKVSARS-COV-OC43FlavivirusesSARS-COV-2DengueZika |
| Drug | N.A. |
| Main interacting RNAs | mRNA |
| Moonlighting functions | E3 ligase |
| Localizations | Stress granucle |
| BulkPerturb-seq | DataSet_02_108 DataSet_02_109 DataSet_02_110 DataSet_02_111 |
Description
| Ensembl ID | ENSG00000121060 | Gene ID | 7706 | Accession | 12932 |
| Symbol | TRIM25 | Alias | EFP;Z147;RNF147;ZNF147 | Full Name | tripartite motif containing 25 |
| Status | Confidence | Length | 77694 bases | Strand | Minus strand |
| Position | 17 : 56836387 - 56914080 | RNA binding domain | SPRY | ||
| Summary | The protein encoded by this gene is a member of the tripartite motif (TRIM) family. The TRIM motif includes three zinc-binding domains, a RING, a B-box type 1 and a B-box type 2, and a coiled-coil region. The protein is an RNA binding protein, functions as a ubiquitin E3 ligase and is involved in multiple cellular processes, including regulation of antiviral innate immunity. [provided by RefSeq, Sep 2021] | ||||
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|---|---|---|---|
| 35800389 | The Role of ZAP and TRIM25 RNA Binding in Restricting Viral Translation. | Emily Yang | 2022-01-01 | Frontiers in cellular and infection microbiology |
| 29117863 | RNA-binding activity of TRIM25 is mediated by its PRY/SPRY domain and is required for ubiquitination. | Roy Nila Choudhury | 2017-11-08 | BMC biology |
| 36171622 | Long non-coding RNA NEAT1 mediated RPRD1B stability facilitates fatty acid metabolism and lymph node metastasis via c-Jun/c-Fos/SREBP1 axis in gastric cancer. | Yongxu Jia | 2022-09-29 | Journal of experimental & clinical cancer research : CR |
| 26138103 | Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. | Gayathri Manokaran | 2015-10-09 | Science (New York, N.Y.) |
| 31417727 | Disease classification via gene network integrating modules and pathways. | Zhilong Mi | 2019-07-01 | Royal Society open science |
| 34781999 | CircRNA-DOPEY2 enhances the chemosensitivity of esophageal cancer cells by inhibiting CPEB4-mediated Mcl-1 translation. | Zhenchuan Liu | 2021-11-15 | Journal of experimental & clinical cancer research : CR |
| 30342007 | TRIM25 Binds RNA to Modulate Cellular Anti-viral Defense. | G Jacint Sanchez | 2018-12-07 | Journal of molecular biology |
| 31754723 | A double-stranded RNA platform is required for the interaction between a host restriction factor and the NS1 protein of influenza A virus. | Guifang Chen | 2020-01-10 | Nucleic acids research |
| 34662337 | Network potential identifies therapeutic miRNA cocktails in Ewing sarcoma. | T Davis Weaver | 2021-10-01 | PLoS computational biology |
| 34145268 | Subtype-specific collaborative transcription factor networks are promoted by OCT4 in the progression of prostate cancer. | Ken-Ichi Takayama | 2021-06-18 | Nature communications |
| 28576494 | Antagonism of type I interferon by flaviviruses. | Lisa Miorin | 2017-10-28 | Biochemical and biophysical research communications |
| 22440909 | Cytosolic surveillance and antiviral immunity. | K A Vijay Rathinam | 2011-12-01 | Current opinion in virology |
| 25457611 | Trim25 Is an RNA-Specific Activator of Lin28a/TuT4-Mediated Uridylation. | Roy Nila Choudhury | 2014-11-20 | Cell reports |
| 33436560 | circNDUFB2 inhibits non-small cell lung cancer progression via destabilizing IGF2BPs and activating anti-tumor immunity. | Botai Li | 2021-01-12 | Nature communications |
| 35533151 | TRIM25 and ZAP target the Ebola virus ribonucleoprotein complex to mediate interferon-induced restriction. | Pedro Rui Galão | 2022-05-01 | PLoS pathogens |
| 33330133 | Regulation of Host Innate Immunity by Non-Coding RNAs During Dengue Virus Infection. | Roopali Rajput | 2020-01-01 | Frontiers in cellular and infection microbiology |
| 31842382 | Identification of TRIM25 as a Negative Regulator of Caspase-2 Expression Reveals a Novel Target for Sensitizing Colon Carcinoma Cells to Intrinsic Apoptosis. | Usman Nasrullah | 2019-12-12 | Cells |
| 29107643 | Nuclear TRIM25 Specifically Targets Influenza Virus Ribonucleoproteins to Block the Onset of RNA Chain Elongation. | R Nicholas Meyerson | 2017-11-08 | Cell host & microbe |
| 35736141 | TRIM25 inhibits influenza A virus infection, destabilizes viral mRNA, but is redundant for activating the RIG-I pathway. | Roy Nila Choudhury | 2022-07-08 | Nucleic acids research |
| 33901262 | Origin and evolution of the zinc finger antiviral protein. | Daniel Gonçalves-Carneiro | 2021-04-01 | PLoS pathogens |
| 26687707 | Phosphorylation of influenza A virus NS1 protein at threonine 49 suppresses its interferon antagonistic activity. | Abid Omer Kathum | 2016-06-01 | Cellular microbiology |
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| TRIM25-210 | ENST00000648772 | 4248 | 456aa | ENSP00000498158 |
| TRIM25-212 | ENST00000682766 | 3366 | 456aa | ENSP00000507876 |
| TRIM25-201 | ENST00000316881 | 5745 | 630aa | ENSP00000323889 |
| TRIM25-207 | ENST00000573108 | 952 | No protein | - |
| TRIM25-202 | ENST00000537230 | 2915 | 630aa | ENSP00000445961 |
| TRIM25-205 | ENST00000572021 | 2132 | 215aa | ENSP00000459980 |
| TRIM25-208 | ENST00000574234 | 471 | No protein | - |
| TRIM25-203 | ENST00000570473 | 498 | No protein | - |
| TRIM25-204 | ENST00000570749 | 614 | No protein | - |
| TRIM25-211 | ENST00000682509 | 1663 | No protein | - |
| TRIM25-206 | ENST00000572550 | 519 | No protein | - |
| TRIM25-209 | ENST00000574997 | 440 | No protein | - |
Phenotypes
| ensgID | Trait | pValue | Pubmed ID |
|---|
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
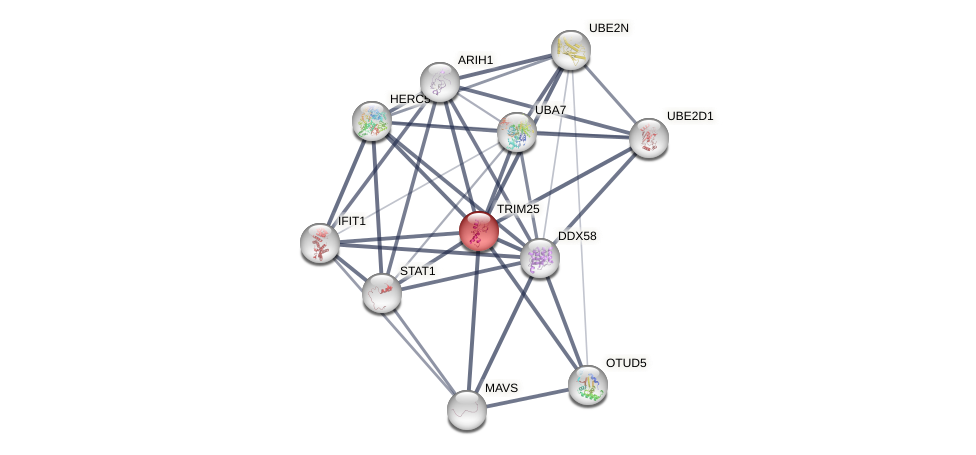
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000121060 | homo_sapiens | ENSP00000366404 | 32.5536 | 20.6628 | homo_sapiens | ENSP00000323889 | 26.5079 | 16.8254 |
| ENSG00000121060 | homo_sapiens | ENSP00000256649 | 25.8621 | 14.8276 | homo_sapiens | ENSP00000323889 | 23.8095 | 13.6508 |
| ENSG00000121060 | homo_sapiens | ENSP00000254816 | 22.4138 | 15.3605 | homo_sapiens | ENSP00000323889 | 22.6984 | 15.5556 |
| ENSG00000121060 | homo_sapiens | ENSP00000496301 | 27.5862 | 17.2414 | homo_sapiens | ENSP00000323889 | 24.127 | 15.0794 |
| ENSG00000121060 | homo_sapiens | ENSP00000518164 | 32.0594 | 20.5945 | homo_sapiens | ENSP00000323889 | 23.9683 | 15.3968 |
| ENSG00000121060 | homo_sapiens | ENSP00000421142 | 33.3333 | 20.8068 | homo_sapiens | ENSP00000323889 | 24.9206 | 15.5556 |
| ENSG00000121060 | homo_sapiens | ENSP00000423934 | 20.5307 | 11.8715 | homo_sapiens | ENSP00000323889 | 23.3333 | 13.4921 |
| ENSG00000121060 | homo_sapiens | ENSP00000365884 | 33.9785 | 20.2151 | homo_sapiens | ENSP00000323889 | 25.0794 | 14.9206 |
| ENSG00000121060 | homo_sapiens | ENSP00000351250 | 13.3097 | 6.92103 | homo_sapiens | ENSP00000323889 | 23.8095 | 12.381 |
| ENSG00000121060 | homo_sapiens | ENSP00000262294 | 14.4191 | 7.67635 | homo_sapiens | ENSP00000323889 | 22.0635 | 11.746 |
| ENSG00000121060 | homo_sapiens | ENSP00000327994 | 33.2649 | 21.1499 | homo_sapiens | ENSP00000323889 | 25.7143 | 16.3492 |
| ENSG00000121060 | homo_sapiens | ENSP00000254436 | 36.8421 | 22.3158 | homo_sapiens | ENSP00000323889 | 27.7778 | 16.8254 |
| ENSG00000121060 | homo_sapiens | ENSP00000343129 | 29.0816 | 16.8367 | homo_sapiens | ENSP00000323889 | 27.1429 | 15.7143 |
| ENSG00000121060 | homo_sapiens | ENSP00000507131 | 12.7044 | 6.41509 | homo_sapiens | ENSP00000323889 | 16.0317 | 8.09524 |
| ENSG00000121060 | homo_sapiens | ENSP00000355613 | 12.1328 | 6.64112 | homo_sapiens | ENSP00000323889 | 15.0794 | 8.25397 |
| ENSG00000121060 | homo_sapiens | ENSP00000474678 | 28.3186 | 15.708 | homo_sapiens | ENSP00000323889 | 20.3175 | 11.2698 |
| ENSG00000121060 | homo_sapiens | ENSP00000485097 | 28.3186 | 15.708 | homo_sapiens | ENSP00000323889 | 20.3175 | 11.2698 |
| ENSG00000121060 | homo_sapiens | ENSP00000402414 | 28.5714 | 16.0714 | homo_sapiens | ENSP00000323889 | 10.1587 | 5.71429 |
| ENSG00000121060 | homo_sapiens | ENSP00000291416 | 33.0526 | 19.5789 | homo_sapiens | ENSP00000323889 | 24.9206 | 14.7619 |
| ENSG00000121060 | homo_sapiens | ENSP00000369373 | 34.8884 | 18.2556 | homo_sapiens | ENSP00000323889 | 27.3016 | 14.2857 |
| ENSG00000121060 | homo_sapiens | ENSP00000268058 | 16.4399 | 9.41043 | homo_sapiens | ENSP00000323889 | 23.0159 | 13.1746 |
| ENSG00000121060 | homo_sapiens | ENSP00000318615 | 31.2 | 19.2 | homo_sapiens | ENSP00000323889 | 12.381 | 7.61905 |
| ENSG00000121060 | homo_sapiens | ENSP00000285805 | 31.2 | 19.2 | homo_sapiens | ENSP00000323889 | 12.381 | 7.61905 |
| ENSG00000121060 | homo_sapiens | ENSP00000474003 | 26.4444 | 16 | homo_sapiens | ENSP00000323889 | 18.8889 | 11.4286 |
| ENSG00000121060 | homo_sapiens | ENSP00000305161 | 15.894 | 8.47682 | homo_sapiens | ENSP00000323889 | 19.0476 | 10.1587 |
| ENSG00000121060 | homo_sapiens | ENSP00000312675 | 28.5115 | 17.1908 | homo_sapiens | ENSP00000323889 | 21.5873 | 13.0159 |
| ENSG00000121060 | homo_sapiens | ENSP00000299413 | 16.8605 | 9.59302 | homo_sapiens | ENSP00000323889 | 9.20635 | 5.2381 |
| ENSG00000121060 | homo_sapiens | ENSP00000349596 | 33.9785 | 21.0753 | homo_sapiens | ENSP00000323889 | 25.0794 | 15.5556 |
| ENSG00000121060 | homo_sapiens | ENSP00000284551 | 34.188 | 21.5812 | homo_sapiens | ENSP00000323889 | 25.3968 | 16.0317 |
| ENSG00000121060 | homo_sapiens | ENSP00000367424 | 31.4496 | 19.1646 | homo_sapiens | ENSP00000323889 | 20.3175 | 12.381 |
| ENSG00000121060 | homo_sapiens | ENSP00000402595 | 32.377 | 18.0328 | homo_sapiens | ENSP00000323889 | 25.0794 | 13.9683 |
| ENSG00000121060 | homo_sapiens | ENSP00000495413 | 8.54885 | 4.31034 | homo_sapiens | ENSP00000323889 | 18.8889 | 9.52381 |
| ENSG00000121060 | homo_sapiens | ENSP00000340797 | 18.9516 | 10.7527 | homo_sapiens | ENSP00000323889 | 22.381 | 12.6984 |
| ENSG00000121060 | homo_sapiens | ENSP00000492719 | 29.148 | 17.4888 | homo_sapiens | ENSP00000323889 | 20.6349 | 12.381 |
| ENSG00000121060 | homo_sapiens | ENSP00000272395 | 28.9238 | 17.713 | homo_sapiens | ENSP00000323889 | 20.4762 | 12.5397 |
| ENSG00000121060 | homo_sapiens | ENSP00000340507 | 14.0952 | 7.2381 | homo_sapiens | ENSP00000323889 | 23.4921 | 12.0635 |
| ENSG00000121060 | homo_sapiens | ENSP00000379800 | 34.6311 | 24.1803 | homo_sapiens | ENSP00000323889 | 26.8254 | 18.7302 |
| ENSG00000121060 | homo_sapiens | ENSP00000481815 | 28.6667 | 17.5556 | homo_sapiens | ENSP00000323889 | 20.4762 | 12.5397 |
| ENSG00000121060 | homo_sapiens | ENSP00000332969 | 29.6214 | 17.8174 | homo_sapiens | ENSP00000323889 | 21.1111 | 12.6984 |
| ENSG00000121060 | homo_sapiens | ENSP00000483764 | 29.8441 | 17.8174 | homo_sapiens | ENSP00000323889 | 21.2698 | 12.6984 |
| ENSG00000121060 | homo_sapiens | ENSP00000332284 | 31.2 | 19.8 | homo_sapiens | ENSP00000323889 | 24.7619 | 15.7143 |
| ENSG00000121060 | homo_sapiens | ENSP00000244360 | 35.2273 | 23.2955 | homo_sapiens | ENSP00000323889 | 19.6825 | 13.0159 |
| ENSG00000121060 | homo_sapiens | ENSP00000491320 | 35.4701 | 20.2991 | homo_sapiens | ENSP00000323889 | 26.3492 | 15.0794 |
| ENSG00000121060 | homo_sapiens | ENSP00000323913 | 21.3504 | 11.8613 | homo_sapiens | ENSP00000323889 | 18.5714 | 10.3175 |
| ENSG00000121060 | homo_sapiens | ENSP00000369415 | 30.1676 | 15.9218 | homo_sapiens | ENSP00000323889 | 17.1429 | 9.04762 |
| ENSG00000121060 | homo_sapiens | ENSP00000363298 | 26.3682 | 14.1791 | homo_sapiens | ENSP00000323889 | 16.8254 | 9.04762 |
| ENSG00000121060 | homo_sapiens | ENSP00000379826 | 31.7829 | 18.9922 | homo_sapiens | ENSP00000323889 | 13.0159 | 7.77778 |
| ENSG00000121060 | homo_sapiens | ENSP00000320869 | 26.6667 | 16.1905 | homo_sapiens | ENSP00000323889 | 26.6667 | 16.1905 |
| ENSG00000121060 | homo_sapiens | ENSP00000508553 | 21.9512 | 15.6794 | homo_sapiens | ENSP00000323889 | 10 | 7.14286 |
| ENSG00000121060 | homo_sapiens | ENSP00000408292 | 22.8178 | 13.6294 | homo_sapiens | ENSP00000323889 | 23.6508 | 14.127 |
| ENSG00000121060 | homo_sapiens | ENSP00000262843 | 26.8027 | 15.5102 | homo_sapiens | ENSP00000323889 | 31.2698 | 18.0952 |
| ENSG00000121060 | homo_sapiens | ENSP00000344208 | 29.8643 | 17.8733 | homo_sapiens | ENSP00000323889 | 20.9524 | 12.5397 |
| ENSG00000121060 | homo_sapiens | ENSP00000373272 | 14.0553 | 8.06452 | homo_sapiens | ENSP00000323889 | 19.3651 | 11.1111 |
| ENSG00000121060 | homo_sapiens | ENSP00000397073 | 30.9771 | 20.5821 | homo_sapiens | ENSP00000323889 | 23.6508 | 15.7143 |
| ENSG00000121060 | homo_sapiens | ENSP00000497185 | 26.9504 | 16.4894 | homo_sapiens | ENSP00000323889 | 24.127 | 14.7619 |
| ENSG00000121060 | homo_sapiens | ENSP00000286349 | 22.4066 | 11.7566 | homo_sapiens | ENSP00000323889 | 25.7143 | 13.4921 |
| ENSG00000121060 | homo_sapiens | ENSP00000339659 | 18.1582 | 10.5058 | homo_sapiens | ENSP00000323889 | 22.2222 | 12.8571 |
| ENSG00000121060 | homo_sapiens | ENSP00000355659 | 34.1719 | 19.7065 | homo_sapiens | ENSP00000323889 | 25.873 | 14.9206 |
| ENSG00000121060 | homo_sapiens | ENSP00000253024 | 14.3713 | 7.54491 | homo_sapiens | ENSP00000323889 | 19.0476 | 10 |
| ENSG00000121060 | homo_sapiens | ENSP00000274773 | 31.3112 | 19.9609 | homo_sapiens | ENSP00000323889 | 25.3968 | 16.1905 |
| ENSG00000121060 | homo_sapiens | ENSP00000507413 | 28.3753 | 18.0778 | homo_sapiens | ENSP00000323889 | 19.6825 | 12.5397 |
| ENSG00000121060 | homo_sapiens | ENSP00000410446 | 26.9017 | 16.8831 | homo_sapiens | ENSP00000323889 | 23.0159 | 14.4444 |
| ENSG00000121060 | homo_sapiens | ENSP00000301924 | 32.2515 | 19.8783 | homo_sapiens | ENSP00000323889 | 25.2381 | 15.5556 |
| ENSG00000121060 | homo_sapiens | ENSP00000363390 | 31.1615 | 18.4136 | homo_sapiens | ENSP00000323889 | 17.4603 | 10.3175 |
| ENSG00000121060 | homo_sapiens | ENSP00000369299 | 34.3373 | 18.4739 | homo_sapiens | ENSP00000323889 | 27.1429 | 14.6032 |
| ENSG00000121060 | homo_sapiens | ENSP00000312678 | 30.2849 | 17.6912 | homo_sapiens | ENSP00000323889 | 32.0635 | 18.7302 |
| ENSG00000121060 | homo_sapiens | ENSP00000327738 | 33.547 | 21.1538 | homo_sapiens | ENSP00000323889 | 24.9206 | 15.7143 |
| ENSG00000121060 | homo_sapiens | ENSP00000330216 | 28.9823 | 16.3717 | homo_sapiens | ENSP00000323889 | 20.7937 | 11.746 |
| ENSG00000121060 | homo_sapiens | ENSP00000388299 | 28.7611 | 16.3717 | homo_sapiens | ENSP00000323889 | 20.6349 | 11.746 |
| ENSG00000121060 | homo_sapiens | ENSP00000327604 | 28.7611 | 16.3717 | homo_sapiens | ENSP00000323889 | 20.6349 | 11.746 |
| ENSG00000121060 | homo_sapiens | ENSP00000269383 | 21.8569 | 14.5068 | homo_sapiens | ENSP00000323889 | 17.9365 | 11.9048 |
| ENSG00000121060 | homo_sapiens | ENSP00000300747 | 34.6392 | 20.8247 | homo_sapiens | ENSP00000323889 | 26.6667 | 16.0317 |
| ENSG00000121060 | homo_sapiens | ENSP00000395086 | 29.646 | 15.9292 | homo_sapiens | ENSP00000323889 | 21.2698 | 11.4286 |
| ENSG00000121060 | homo_sapiens | ENSP00000497050 | 27.8761 | 15.4867 | homo_sapiens | ENSP00000323889 | 20 | 11.1111 |
| ENSG00000121060 | homo_sapiens | ENSP00000275736 | 30.8017 | 20.0422 | homo_sapiens | ENSP00000323889 | 23.1746 | 15.0794 |
| ENSG00000121060 | homo_sapiens | ENSP00000334657 | 18.7088 | 10.9354 | homo_sapiens | ENSP00000323889 | 22.5397 | 13.1746 |
| ENSG00000121060 | homo_sapiens | ENSP00000355437 | 34.5679 | 19.5473 | homo_sapiens | ENSP00000323889 | 26.6667 | 15.0794 |
| ENSG00000121060 | homo_sapiens | ENSP00000365924 | 25.1765 | 14.8235 | homo_sapiens | ENSP00000323889 | 16.9841 | 10 |
| ENSG00000121060 | homo_sapiens | ENSP00000219596 | 13.9565 | 6.40205 | homo_sapiens | ENSP00000323889 | 17.3016 | 7.93651 |
| ENSG00000121060 | homo_sapiens | ENSP00000369440 | 33.5271 | 18.2171 | homo_sapiens | ENSP00000323889 | 27.4603 | 14.9206 |
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 97.4603 | 96.5079 | nomascus_leucogenys | ENSNLEP00000044600 | 97.4603 | 96.5079 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 99.2064 | 98.7302 | pan_paniscus | ENSPPAP00000018579 | 99.2064 | 98.7302 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 97.4603 | 96.0317 | pongo_abelii | ENSPPYP00000009268 | 97.4603 | 96.0317 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 99.2064 | 98.7302 | pan_troglodytes | ENSPTRP00000016020 | 99.2064 | 98.7302 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 99.0476 | 98.5714 | gorilla_gorilla | ENSGGOP00000011502 | 99.0476 | 98.5714 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 70.6349 | 57.3016 | sarcophilus_harrisii | ENSSHAP00000002459 | 65.9259 | 53.4815 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 44.2857 | 38.0952 | notamacropus_eugenii | ENSMEUP00000013678 | 44.1456 | 37.9747 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 30.4762 | 25.3968 | choloepus_hoffmanni | ENSCHOP00000002244 | 53.0387 | 44.1989 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 83.8095 | 74.9206 | dasypus_novemcinctus | ENSDNOP00000028823 | 83.6767 | 74.8019 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 76.5079 | 67.619 | erinaceus_europaeus | ENSEEUP00000012940 | 75.9055 | 67.0866 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 71.5873 | 63.4921 | echinops_telfairi | ENSETEP00000003025 | 79.1228 | 70.1754 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 91.4286 | 86.6667 | callithrix_jacchus | ENSCJAP00000081766 | 89.5801 | 84.9145 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 96.5079 | 94.127 | cercocebus_atys | ENSCATP00000036176 | 96.5079 | 94.127 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 96.9841 | 94.9206 | macaca_fascicularis | ENSMFAP00000062445 | 96.9841 | 94.9206 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 74.2857 | 66.3492 | macaca_mulatta | ENSMMUP00000080547 | 52.0578 | 46.4961 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 96.9841 | 94.9206 | macaca_nemestrina | ENSMNEP00000043042 | 96.9841 | 94.9206 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 96.6667 | 94.4444 | papio_anubis | ENSPANP00000020334 | 96.6667 | 94.4444 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 96.6667 | 94.127 | mandrillus_leucophaeus | ENSMLEP00000017854 | 96.6667 | 94.127 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 62.0635 | 56.6667 | tursiops_truncatus | ENSTTRP00000007665 | 82.3158 | 75.1579 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 69.8413 | 62.381 | vulpes_vulpes | ENSVVUP00000015911 | 84.7784 | 75.7225 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 82.2222 | 72.381 | mus_spretus | MGP_SPRETEiJ_P0030562 | 81.7035 | 71.9243 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 67.4603 | 57.3016 | equus_asinus | ENSEASP00005051505 | 67.2468 | 57.1203 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 80 | 70.6349 | equus_asinus | ENSEASP00005040477 | 81.1594 | 71.6586 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 86.1905 | 76.3492 | capra_hircus | ENSCHIP00000009746 | 85.782 | 75.9874 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 49.3651 | 44.7619 | loxodonta_africana | ENSLAFP00000016399 | 84.5109 | 76.6304 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 81.4286 | 73.8095 | panthera_leo | ENSPLOP00000020318 | 84.375 | 76.4803 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 86.0317 | 78.5714 | ailuropoda_melanoleuca | ENSAMEP00000030757 | 85.489 | 78.0757 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 85.7143 | 76.3492 | bos_indicus_hybrid | ENSBIXP00005024984 | 85.5784 | 76.2282 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 49.2063 | 43.1746 | oryctolagus_cuniculus | ENSOCUP00000009756 | 84.2391 | 73.913 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 79.8413 | 71.4286 | equus_caballus | ENSECAP00000021893 | 81.2601 | 72.6979 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 86.6667 | 78.5714 | equus_caballus | ENSECAP00000020737 | 85.7143 | 77.708 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 84.7619 | 76.1905 | canis_lupus_familiaris | ENSCAFP00845025140 | 84.6276 | 76.0697 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 81.4286 | 73.8095 | panthera_pardus | ENSPPRP00000001637 | 84.375 | 76.4803 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 82.8571 | 70.7936 | marmota_marmota_marmota | ENSMMMP00000023752 | 80.9302 | 69.1473 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 86.8254 | 79.5238 | balaenoptera_musculus | ENSBMSP00010027578 | 86.4139 | 79.1469 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 79.5238 | 69.2064 | cavia_porcellus | ENSCPOP00000012112 | 80.16 | 69.76 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 81.5873 | 74.2857 | felis_catus | ENSFCAP00000007439 | 84.5395 | 76.9737 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 72.5397 | 59.2063 | monodelphis_domestica | ENSMODP00000018527 | 68.006 | 55.506 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 70.7936 | 59.6825 | bison_bison_bison | ENSBBBP00000002017 | 74.8322 | 63.0872 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 86.6667 | 79.0476 | monodon_monoceros | ENSMMNP00015021128 | 86.1199 | 78.5489 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 84.6032 | 76.6667 | sus_scrofa | ENSSSCP00000067922 | 84.2022 | 76.3033 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 88.4127 | 82.0635 | microcebus_murinus | ENSMICP00000028116 | 87.8549 | 81.5457 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 78.7302 | 68.7302 | octodon_degus | ENSODEP00000016071 | 83.3613 | 72.7731 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 80 | 70 | jaculus_jaculus | ENSJJAP00000021275 | 82.4877 | 72.1768 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 79.5238 | 68.4127 | ovis_aries_rambouillet | ENSOARP00020029289 | 68.7243 | 59.1221 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 67.3016 | 59.3651 | ursus_maritimus | ENSUMAP00000000943 | 86.5306 | 76.3265 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 52.2222 | 43.4921 | ochotona_princeps | ENSOPRP00000015017 | 74.0991 | 61.7117 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 86.8254 | 79.2064 | delphinapterus_leucas | ENSDLEP00000008907 | 86.2776 | 78.7066 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 87.1429 | 79.6825 | physeter_catodon | ENSPCTP00005017093 | 81.3333 | 74.3704 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 82.5397 | 72.2222 | mus_caroli | MGP_CAROLIEiJ_P0029148 | 81.8898 | 71.6535 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 82.381 | 73.0159 | mus_musculus | ENSMUSP00000103528 | 81.8612 | 72.5552 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 78.4127 | 67.3016 | dipodomys_ordii | ENSDORP00000023881 | 82.6087 | 70.903 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 85.873 | 77.9365 | mustela_putorius_furo | ENSMPUP00000015278 | 82.8484 | 75.1914 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 91.1111 | 86.8254 | aotus_nancymaae | ENSANAP00000026797 | 92.5806 | 88.2258 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 87.7778 | 83.6508 | saimiri_boliviensis_boliviensis | ENSSBOP00000001608 | 92.7852 | 88.4228 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 45.0794 | 40.4762 | vicugna_pacos | ENSVPAP00000008023 | 65.5889 | 58.8915 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 96.9841 | 94.2857 | chlorocebus_sabaeus | ENSCSAP00000001899 | 96.9841 | 94.2857 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 52.0635 | 45.7143 | tupaia_belangeri | ENSTBEP00000010158 | 82.4121 | 72.3618 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 76.9841 | 67.9365 | mesocricetus_auratus | ENSMAUP00000005537 | 80.5648 | 71.0963 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 86.1905 | 78.5714 | phocoena_sinus | ENSPSNP00000018474 | 85.5118 | 77.9528 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 85.7143 | 77.7778 | camelus_dromedarius | ENSCDRP00005032525 | 85.3081 | 77.4092 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 87.9365 | 82.0635 | carlito_syrichta | ENSTSYP00000014972 | 89.4992 | 83.5218 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 70.3175 | 63.0159 | panthera_tigris_altaica | ENSPTIP00000002892 | 82.6493 | 74.0672 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 82.5397 | 72.6984 | rattus_norvegicus | ENSRNOP00000052533 | 80.8709 | 71.2286 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 88.254 | 82.381 | prolemur_simus | ENSPSMP00000029521 | 88.254 | 82.381 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 82.0635 | 72.8571 | microtus_ochrogaster | ENSMOCP00000004408 | 81.5457 | 72.3975 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 49.5238 | 42.381 | sorex_araneus | ENSSARP00000011150 | 70.1124 | 60 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 82.5397 | 72.5397 | peromyscus_maniculatus_bairdii | ENSPEMP00000026637 | 81.6327 | 71.7425 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 68.5714 | 60 | bos_mutus | ENSBMUP00000033592 | 68.7898 | 60.1911 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 82.2222 | 72.6984 | mus_spicilegus | ENSMSIP00000036921 | 81.7035 | 72.2397 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 82.381 | 72.8571 | cricetulus_griseus_chok1gshd | ENSCGRP00001013500 | 81.7323 | 72.2835 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 87.7778 | 81.1111 | pteropus_vampyrus | ENSPVAP00000003709 | 87.5 | 80.8544 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 86.6667 | 77.619 | myotis_lucifugus | ENSMLUP00000009789 | 86.2559 | 77.2512 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 22.0635 | 17.1429 | vombatus_ursinus | ENSVURP00010000100 | 72.7749 | 56.5445 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 54.6032 | 39.6825 | vombatus_ursinus | ENSVURP00010017772 | 35.6108 | 25.8799 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 69.3651 | 55.3968 | vombatus_ursinus | ENSVURP00010017761 | 68.6028 | 54.7881 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 87.9365 | 80 | rhinolophus_ferrumequinum | ENSRFEP00010025329 | 87.3817 | 79.4953 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 85.7143 | 78.0952 | neovison_vison | ENSNVIP00000015685 | 84.9057 | 77.3585 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 85.7143 | 76.3492 | bos_taurus | ENSBTAP00000013121 | 85.5784 | 76.2282 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 81.5873 | 70.3175 | heterocephalus_glaber_female | ENSHGLP00000016047 | 81.8471 | 70.5414 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 96.9841 | 94.4444 | rhinopithecus_bieti | ENSRBIP00000038618 | 96.9841 | 94.4444 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 84.7619 | 76.1905 | canis_lupus_dingo | ENSCAFP00020032967 | 84.6276 | 76.0697 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 81.9048 | 71.4286 | sciurus_vulgaris | ENSSVLP00005011373 | 83.0918 | 72.4638 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 70.9524 | 55.3968 | phascolarctos_cinereus | ENSPCIP00000047204 | 66.7164 | 52.0896 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 37.9365 | 27.3016 | phascolarctos_cinereus | ENSPCIP00000054534 | 63.9037 | 45.9893 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 82.6984 | 73.9683 | procavia_capensis | ENSPCAP00000004785 | 83.2268 | 74.4409 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 80.9524 | 70.7936 | bos_grunniens | ENSBGRP00000027451 | 70.5394 | 61.6874 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 82.0635 | 70.9524 | chinchilla_lanigera | ENSCLAP00000024812 | 82.194 | 71.0652 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 85.3968 | 76.5079 | cervus_hanglu_yarkandensis | ENSCHYP00000020061 | 84.9921 | 76.1453 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 83.3333 | 74.6032 | catagonus_wagneri | ENSCWAP00000011202 | 82.5472 | 73.8994 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 82.381 | 71.1111 | ictidomys_tridecemlineatus | ENSSTOP00000008370 | 80.5901 | 69.5652 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 82.6984 | 75.2381 | otolemur_garnettii | ENSOGAP00000003441 | 83.0941 | 75.5981 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 89.3651 | 84.7619 | cebus_imitator | ENSCCAP00000021590 | 90.8064 | 86.129 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 82.0635 | 70.1587 | urocitellus_parryii | ENSUPAP00010019014 | 81.4173 | 69.6063 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 86.3492 | 77.3016 | moschus_moschiferus | ENSMMSP00000017990 | 85.94 | 76.9352 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 96.9841 | 94.2857 | rhinopithecus_roxellana | ENSRROP00000026382 | 96.9841 | 94.2857 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 82.2222 | 71.5873 | mus_pahari | MGP_PahariEiJ_P0035862 | 81.5748 | 71.0236 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 88.5714 | 83.4921 | propithecus_coquereli | ENSPCOP00000026732 | 88.1517 | 83.0964 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 55.3968 | 37.7778 | latimeria_chalumnae | ENSLACP00000006406 | 53.1202 | 36.2253 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 54.9206 | 38.8889 | xenopus_tropicalis | ENSXETP00000105309 | 54.232 | 38.4013 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 66.1905 | 50.6349 | chrysemys_picta_bellii | ENSCPBP00000005689 | 65.6693 | 50.2362 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 64.2857 | 48.7302 | sphenodon_punctatus | ENSSPUP00000006963 | 63.981 | 48.4992 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 65.5556 | 49.2063 | crocodylus_porosus | ENSCPRP00005002863 | 64.5312 | 48.4375 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 43.1746 | 29.0476 | laticauda_laticaudata | ENSLLTP00000008034 | 61.678 | 41.4966 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 59.0476 | 43.9683 | notechis_scutatus | ENSNSUP00000002332 | 61.6915 | 45.937 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 59.5238 | 44.2857 | pseudonaja_textilis | ENSPTXP00000009488 | 61.9835 | 46.1157 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 65.5556 | 49.2063 | anas_platyrhynchos_platyrhynchos | ENSAPLP00000018873 | 63.7346 | 47.8395 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 64.4444 | 47.1429 | gallus_gallus | ENSGALP00010044203 | 60.4167 | 44.1964 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 63.8095 | 47.619 | meleagris_gallopavo | ENSMGAP00000023311 | 61.9414 | 46.225 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 59.6825 | 43.9683 | serinus_canaria | ENSSCAP00000019857 | 60.2564 | 44.391 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 61.2698 | 45.5556 | parus_major | ENSPMJP00000024625 | 61.465 | 45.7006 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 63.4921 | 48.0952 | anolis_carolinensis | ENSACAP00000009034 | 62.3053 | 47.1963 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 56.5079 | 42.2222 | naja_naja | ENSNNAP00000005244 | 61.913 | 46.2609 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 35.5556 | 29.5238 | podarcis_muralis | ENSPMRP00000009093 | 60.8696 | 50.5435 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 49.6825 | 35.7143 | geospiza_fortis | ENSGFOP00000014973 | 63.4888 | 45.6389 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 62.0635 | 46.0317 | taeniopygia_guttata | ENSTGUP00000022248 | 61.2853 | 45.4545 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 63.1746 | 48.254 | salvator_merianae | ENSSMRP00000028145 | 61.8012 | 47.205 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 66.5079 | 51.746 | pelodiscus_sinensis | ENSPSIP00000009688 | 61.7994 | 48.0826 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 66.5079 | 51.1111 | gopherus_evgoodei | ENSGEVP00005018251 | 65.9843 | 50.7087 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 66.5079 | 51.4286 | chelonoidis_abingdonii | ENSCABP00000006912 | 67.2552 | 52.0064 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 54.7619 | 39.8413 | struthio_camelus_australis | ENSSCUP00000015705 | 67.5147 | 49.1194 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 63.9683 | 49.2063 | anser_brachyrhynchus | ENSABRP00000014190 | 60.2392 | 46.3378 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 60.7937 | 43.6508 | strigops_habroptila | ENSSHBP00005025213 | 61.3782 | 44.0705 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 63.1746 | 47.4603 | terrapene_carolina_triunguis | ENSTMTP00000012066 | 68.1507 | 51.1986 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 63.8095 | 47.1429 | aquila_chrysaetos_chrysaetos | ENSACCP00020008352 | 63.0094 | 46.5517 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 61.746 | 45.3968 | ficedula_albicollis | ENSFALP00000002369 | 56.623 | 41.6303 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 64.7619 | 47.1429 | coturnix_japonica | ENSCJPP00005027645 | 63.9498 | 46.5517 |
| ENSG00000121060 | homo_sapiens | ENSP00000323889 | 55.873 | 38.8889 | leptobrachium_leishanense | ENSLLEP00000049332 | 50.5747 | 35.2011 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0002753 | involved_in cytosolic pattern recognition receptor signaling pathway | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0003713 | enables transcription coactivator activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0003723 | enables RNA binding | 1 | HDA | Homo_sapiens(9606) | Function |
| GO:0004842 | enables ubiquitin-protein transferase activity | 1 | EXP | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005654 | located_in nucleoplasm | 2 | IDA,TAS | Homo_sapiens(9606) | Component |
| GO:0005737 | is_active_in cytoplasm | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005829 | located_in cytosol | 2 | IDA,TAS | Homo_sapiens(9606) | Component |
| GO:0006511 | involved_in ubiquitin-dependent protein catabolic process | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0006513 | involved_in protein monoubiquitination | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0006979 | involved_in response to oxidative stress | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0010494 | located_in cytoplasmic stress granule | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0016604 | located_in nuclear body | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0016874 | enables ligase activity | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0019076 | involved_in viral release from host cell | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0030433 | involved_in ubiquitin-dependent ERAD pathway | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0032880 | involved_in regulation of protein localization | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0033280 | involved_in response to vitamin D | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0039529 | NOT involved_in RIG-I signaling pathway | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0039552 | enables RIG-I binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0043123 | involved_in positive regulation of I-kappaB kinase/NF-kappaB signaling | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0043627 | involved_in response to estrogen | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0044790 | involved_in suppression of viral release by host | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0045087 | involved_in innate immune response | 2 | IDA,TAS | Homo_sapiens(9606) | Process |
| GO:0045296 | enables cadherin binding | 1 | HDA | Homo_sapiens(9606) | Function |
| GO:0045893 | involved_in positive regulation of DNA-templated transcription | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0046596 | involved_in regulation of viral entry into host cell | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0046597 | involved_in negative regulation of viral entry into host cell | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0046872 | enables metal ion binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0051091 | involved_in positive regulation of DNA-binding transcription factor activity | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0051092 | involved_in positive regulation of NF-kappaB transcription factor activity | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0061630 | enables ubiquitin protein ligase activity | 2 | IDA,IMP | Homo_sapiens(9606) | Function |
| GO:0070936 | involved_in protein K48-linked ubiquitination | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0140374 | involved_in antiviral innate immune response | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:1990830 | involved_in cellular response to leukemia inhibitory factor | 1 | IEA | Homo_sapiens(9606) | Process |