Welcome to
RBP World!
Description
| Ensembl ID | ENSG00000109819 | Gene ID | 10891 | Accession | 9237 |
| Symbol | PPARGC1A | Alias | LEM6;PGC1;PGC1A;PGC-1v;PPARGC1;PGC-1alpha;PGC-1(alpha) | Full Name | PPARG coactivator 1 alpha |
| Status | Confidence | Length | 98057 bases | Strand | Minus strand |
| Position | 4 : 23792021 - 23890077 | RNA binding domain | RRM_1 | ||
| Summary | The protein encoded by this gene is a transcriptional coactivator that regulates the genes involved in energy metabolism. This protein interacts with PPARgamma, which permits the interaction of this protein with multiple transcription factors. This protein can interact with, and regulate the activities of, cAMP response element binding protein (CREB) and nuclear respiratory factors (NRFs). It provides a direct link between external physiological stimuli and the regulation of mitochondrial biogenesis, and is a major factor that regulates muscle fiber type determination. This protein may be also involved in controlling blood pressure, regulating cellular cholesterol homoeostasis, and the development of obesity. [provided by RefSeq, Jul 2008] | ||||
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|---|---|---|---|
| 36153371 | Regulatory chromatin rewiring promotes metabolic switching during adaptation to oncogenic receptor tyrosine kinase inhibition. | Samuel Ogden | 2022-10-01 | Oncogene |
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| PPARGC1A-209 | ENST00000509702 | 2704 | No protein | - |
| PPARGC1A-201 | ENST00000264867 | 6288 | 798aa | ENSP00000264867 |
| PPARGC1A-204 | ENST00000506055 | 2627 | 271aa | ENSP00000423075 |
| PPARGC1A-211 | ENST00000513205 | 1432 | 301aa | ENSP00000421632 |
| PPARGC1A-208 | ENST00000509642 | 1391 | No protein | - |
| PPARGC1A-212 | ENST00000514479 | 324 | No protein | - |
| PPARGC1A-207 | ENST00000508380 | 576 | No protein | - |
| PPARGC1A-210 | ENST00000512169 | 550 | No protein | - |
| PPARGC1A-205 | ENST00000507342 | 359 | No protein | - |
| PPARGC1A-215 | ENST00000515534 | 433 | No protein | - |
| PPARGC1A-202 | ENST00000503714 | 380 | No protein | - |
| PPARGC1A-203 | ENST00000505469 | 406 | No protein | - |
| PPARGC1A-214 | ENST00000514517 | 548 | No protein | - |
| PPARGC1A-206 | ENST00000507380 | 2385 | 99aa | ENSP00000424181 |
| PPARGC1A-213 | ENST00000514494 | 529 | No protein | - |
| PPARGC1A-217 | ENST00000613098 | 3210 | 671aa | ENSP00000481498 |
| PPARGC1A-218 | ENST00000617484 | 1026 | 297aa | ENSP00000477921 |
| PPARGC1A-216 | ENST00000612355 | 936 | 267aa | ENSP00000479729 |
Phenotypes
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| 3470 | Cholesterol, HDL | 9.11565346408683E-5 | 17903299 |
| 4704 | Cholesterol, HDL | 8.92867062780578E-5 | 17903299 |
| 5693 | Blood Pressure | 5.04278146959727E-6 | 17903303 |
| 5696 | Blood Pressure | 8.19596529333388E-6 | 17903303 |
| 5706 | Blood Pressure | 8.58702893994373E-6 | 17903303 |
| 5709 | Blood Pressure | 1.01477958305285E-5 | 17903303 |
| 7189 | Alkaline Phosphatase | 5.09992075077292E-5 | 17903293 |
| 13898 | Myocardial Infarction | 7.3233145E-004 | - |
| 15349 | Heart Failure | 4.0505419E-004 | - |
| 16083 | Stroke | 3.4733485E-004 | 19369658 |
| 17127 | Blood Pressure | 1.137e-005 | 19060910 |
| 21717 | Personality | 3.327e-005 | - |
| 27447 | Cardiovascular Physiological Phenomena | 1.2830000E-005 | - |
| 27448 | Cardiovascular Physiological Phenomena | 4.2550000E-005 | - |
| 27493 | Cardiovascular Physiological Phenomena | 6.5600000E-005 | - |
| 29635 | Lipoprotein(a) | 8.6620000E-005 | - |
| 29636 | Lipoprotein(a) | 4.4610000E-005 | - |
| 29652 | Lipoprotein(a) | 7.7620000E-005 | - |
| 29655 | Lipoprotein(a) | 8.8630000E-005 | - |
| 30535 | Monocytes | 3.4470000E-005 | - |
| 31639 | Platelet Count | 1.2390000E-005 | - |
| 39633 | Platelet Function Tests | 2.8111000E-005 | - |
| 43602 | Platelet Function Tests | 2.3248900E-005 | - |
| 52328 | Child Development Disorders, Pervasive | 4.4000000E-005 | - |
| 56276 | Erythrocyte Count | 6.4500000E-006 | - |
| 70529 | Tooth Demineralization | 2E-6 | 23918034 |
| 73047 | Smell | 9E-6 | 26632684 |
| 77606 | Cognition Disorders | 1E-7 | 26252872 |
| 77607 | Cognitive Dysfunction | 1E-7 | 26252872 |
| 81953 | Schizophrenia | 1E-6 | 26198764 |
| 84819 | Emphysema | 2E-10 | 24383474 |
| 86784 | Gout | 1E-7 | 22179738 |
| 94538 | Cognition Disorders | 7E-8 | 26252872 |
| 94539 | Cognitive Dysfunction | 7E-8 | 26252872 |
| 97071 | Longevity | 6E-6 | 27015805 |
| 101605 | Heart Rate | 2E-15 | 27798624 |
| 105923 | Essential Tremor | 1E-9 | 27797806 |
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
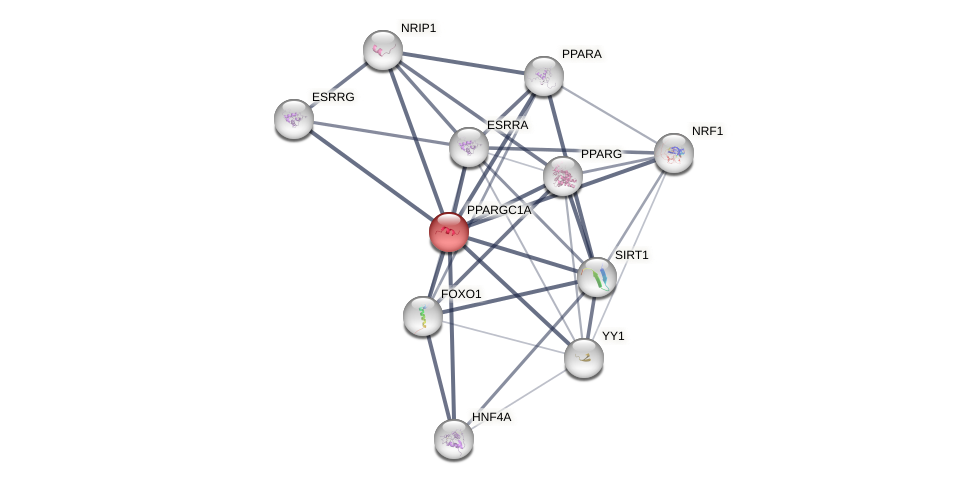
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000109819 | homo_sapiens | ENSP00000312649 | 34.4086 | 24.6334 | homo_sapiens | ENSP00000264867 | 44.1103 | 31.5789 |
| ENSG00000109819 | homo_sapiens | ENSP00000278070 | 19.5312 | 12.5 | homo_sapiens | ENSP00000264867 | 40.7268 | 26.0652 |
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 98.9975 | 98.6216 | nomascus_leucogenys | ENSNLEP00000041737 | 98.6267 | 98.2522 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 99.7494 | 99.6241 | pan_paniscus | ENSPPAP00000023052 | 98.3931 | 98.2695 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 99.8747 | 99.8747 | pongo_abelii | ENSPPYP00000016357 | 99.8747 | 99.8747 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 99.7494 | 99.7494 | pan_troglodytes | ENSPTRP00000027470 | 98.3931 | 98.3931 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 99.7494 | 99.7494 | gorilla_gorilla | ENSGGOP00000009586 | 99.7494 | 99.7494 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 92.7318 | 88.5965 | ornithorhynchus_anatinus | ENSOANP00000043917 | 93.0818 | 88.9308 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 92.8571 | 89.7243 | sarcophilus_harrisii | ENSSHAP00000011681 | 87.2792 | 84.3345 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 61.0276 | 58.6466 | notamacropus_eugenii | ENSMEUP00000005489 | 72.6866 | 69.8507 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 90.8521 | 89.0977 | choloepus_hoffmanni | ENSCHOP00000000325 | 91.195 | 89.434 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 95.99 | 94.6115 | dasypus_novemcinctus | ENSDNOP00000017677 | 95.99 | 94.6115 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 93.8596 | 92.1053 | erinaceus_europaeus | ENSEEUP00000008402 | 96.0256 | 94.2308 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 87.4687 | 83.0827 | echinops_telfairi | ENSETEP00000014701 | 89.3726 | 84.8912 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 99.3734 | 98.9975 | callithrix_jacchus | ENSCJAP00000046572 | 99.3734 | 98.9975 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 99.6241 | 99.6241 | cercocebus_atys | ENSCATP00000017431 | 99.6241 | 99.6241 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 97.4937 | 96.9925 | macaca_fascicularis | ENSMFAP00000045769 | 97.0075 | 96.5087 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 99.4987 | 99.2481 | macaca_mulatta | ENSMMUP00000060440 | 97.066 | 96.8215 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 99.4987 | 99.2481 | macaca_nemestrina | ENSMNEP00000008354 | 99.4987 | 99.2481 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 99.6241 | 99.6241 | papio_anubis | ENSPANP00000018518 | 99.6241 | 99.6241 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 99.6241 | 99.6241 | mandrillus_leucophaeus | ENSMLEP00000020141 | 99.6241 | 99.6241 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 94.8622 | 93.7343 | tursiops_truncatus | ENSTTRP00000005033 | 95.1005 | 93.9698 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 96.9925 | 94.9875 | vulpes_vulpes | ENSVVUP00000018149 | 94.7368 | 92.7785 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 96.8672 | 94.1103 | mus_spretus | MGP_SPRETEiJ_P0070780 | 97.1105 | 94.3467 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 98.1203 | 96.9925 | equus_asinus | ENSEASP00005040346 | 96.5475 | 95.4377 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 96.9925 | 94.8622 | capra_hircus | ENSCHIP00000015380 | 97.1142 | 94.9812 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 96.8672 | 94.8622 | loxodonta_africana | ENSLAFP00000010853 | 97.1105 | 95.1005 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 96.9925 | 95.8647 | panthera_leo | ENSPLOP00000021388 | 97.2362 | 96.1055 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 88.3459 | 87.218 | ailuropoda_melanoleuca | ENSAMEP00000011007 | 97.9167 | 96.6667 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 96.6165 | 94.7368 | bos_indicus_hybrid | ENSBIXP00005011488 | 96.8593 | 94.9749 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 96.4912 | 94.6115 | oryctolagus_cuniculus | ENSOCUP00000034883 | 89.5349 | 87.7907 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 97.2431 | 95.7393 | equus_caballus | ENSECAP00000066433 | 91.8343 | 90.4142 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 96.6165 | 94.9875 | canis_lupus_familiaris | ENSCAFP00845011775 | 89.547 | 88.0372 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 96.9925 | 95.99 | panthera_pardus | ENSPPRP00000006178 | 97.2362 | 96.2312 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 97.4937 | 95.614 | marmota_marmota_marmota | ENSMMMP00000019477 | 97.7387 | 95.8543 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 96.2406 | 94.3609 | balaenoptera_musculus | ENSBMSP00010002551 | 96.4824 | 94.598 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 97.4937 | 95.7393 | cavia_porcellus | ENSCPOP00000022036 | 97.7387 | 95.9799 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 95.614 | 94.4862 | felis_catus | ENSFCAP00000054723 | 95.0187 | 93.8979 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 89.0977 | 87.218 | ursus_americanus | ENSUAMP00000027059 | 93.4297 | 91.4586 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 93.4837 | 90.8521 | monodelphis_domestica | ENSMODP00000000664 | 93.8365 | 91.195 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 95.614 | 93.7343 | bison_bison_bison | ENSBBBP00000021689 | 95.8543 | 93.9698 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 96.1153 | 94.7368 | monodon_monoceros | ENSMMNP00015011962 | 96.3568 | 94.9749 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 96.7419 | 94.7368 | sus_scrofa | ENSSSCP00000032437 | 96.9849 | 94.9749 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 97.8697 | 95.8647 | microcebus_murinus | ENSMICP00000045541 | 93.7575 | 91.8367 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 33.3333 | 32.4561 | octodon_degus | ENSODEP00000011314 | 98.8848 | 96.2825 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 33.4586 | 32.3308 | jaculus_jaculus | ENSJJAP00000016556 | 99.2565 | 95.9108 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 95.8647 | 93.609 | ovis_aries_rambouillet | ENSOARP00020018739 | 86.1487 | 84.1216 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 95.614 | 94.4862 | ursus_maritimus | ENSUMAP00000004473 | 97.3214 | 96.1735 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 96.3659 | 94.3609 | ochotona_princeps | ENSOPRP00000011781 | 96.8514 | 94.8363 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 96.1153 | 94.8622 | delphinapterus_leucas | ENSDLEP00000019612 | 96.3568 | 95.1005 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 96.4912 | 94.6115 | physeter_catodon | ENSPCTP00005016421 | 96.7337 | 94.8492 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 96.8672 | 94.1103 | mus_caroli | MGP_CAROLIEiJ_P0068146 | 97.1105 | 94.3467 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 97.1178 | 94.3609 | mus_musculus | ENSMUSP00000117040 | 97.2396 | 94.4793 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 96.6165 | 93.985 | dipodomys_ordii | ENSDORP00000015313 | 96.7378 | 94.1029 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 96.8672 | 95.2381 | mustela_putorius_furo | ENSMPUP00000016632 | 97.1105 | 95.4774 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 98.3709 | 97.4937 | aotus_nancymaae | ENSANAP00000036857 | 97.8803 | 97.0075 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 97.619 | 97.1178 | saimiri_boliviensis_boliviensis | ENSSBOP00000001315 | 97.1322 | 96.6334 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 88.2206 | 86.0902 | vicugna_pacos | ENSVPAP00000003222 | 88.4422 | 86.3065 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 99.6241 | 99.6241 | chlorocebus_sabaeus | ENSCSAP00000012633 | 99.6241 | 99.6241 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 90.1003 | 88.5965 | tupaia_belangeri | ENSTBEP00000004443 | 90.1003 | 88.5965 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 97.1178 | 94.4862 | mesocricetus_auratus | ENSMAUP00000011137 | 97.3618 | 94.7236 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 96.3659 | 95.1128 | phocoena_sinus | ENSPSNP00000028395 | 96.608 | 95.3518 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 95.3634 | 92.9825 | camelus_dromedarius | ENSCDRP00005023243 | 95.2441 | 92.8661 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 99.3734 | 98.9975 | carlito_syrichta | ENSTSYP00000018015 | 99.3734 | 98.9975 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 96.9925 | 95.99 | panthera_tigris_altaica | ENSPTIP00000005088 | 97.2362 | 96.2312 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 95.7393 | 93.2331 | rattus_norvegicus | ENSRNOP00000093101 | 97.201 | 94.6565 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 98.7469 | 97.8697 | prolemur_simus | ENSPSMP00000029446 | 98.7469 | 97.8697 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 96.7419 | 94.1103 | microtus_ochrogaster | ENSMOCP00000008731 | 96.9849 | 94.3467 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 90.1003 | 88.4712 | sorex_araneus | ENSSARP00000000529 | 90.3266 | 88.6935 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 97.2431 | 94.9875 | peromyscus_maniculatus_bairdii | ENSPEMP00000009669 | 97.3651 | 95.1067 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 96.6165 | 94.7368 | bos_mutus | ENSBMUP00000011587 | 96.8593 | 94.9749 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 96.9925 | 94.4862 | mus_spicilegus | ENSMSIP00000023605 | 95.086 | 92.629 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 96.9925 | 94.6115 | cricetulus_griseus_chok1gshd | ENSCGRP00001014751 | 97.2362 | 94.8492 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 97.995 | 96.3659 | pteropus_vampyrus | ENSPVAP00000004697 | 98.2412 | 96.608 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 97.2431 | 95.614 | myotis_lucifugus | ENSMLUP00000014864 | 97.4874 | 95.8543 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 69.6742 | 66.792 | vombatus_ursinus | ENSVURP00010005581 | 87.011 | 83.4116 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 97.7444 | 96.2406 | rhinolophus_ferrumequinum | ENSRFEP00010002448 | 97.99 | 96.4824 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 96.8672 | 95.3634 | neovison_vison | ENSNVIP00000016625 | 97.1105 | 95.603 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 95.4887 | 93.4837 | bos_taurus | ENSBTAP00000022636 | 93.0403 | 91.0867 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 97.1178 | 95.2381 | heterocephalus_glaber_female | ENSHGLP00000009786 | 96.6334 | 94.7631 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 99.3734 | 99.3734 | rhinopithecus_bieti | ENSRBIP00000016637 | 99.3734 | 99.3734 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 95.1128 | 93.7343 | canis_lupus_dingo | ENSCAFP00020035173 | 98.062 | 96.6408 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 97.7444 | 95.7393 | sciurus_vulgaris | ENSSVLP00005018554 | 97.99 | 95.9799 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 92.8571 | 89.599 | phascolarctos_cinereus | ENSPCIP00000020825 | 93.2076 | 89.9371 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 95.99 | 92.7318 | procavia_capensis | ENSPCAP00000015260 | 96.2312 | 92.9648 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 96.9925 | 93.985 | nannospalax_galili | ENSNGAP00000013544 | 97.2362 | 94.2211 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 96.6165 | 94.7368 | bos_grunniens | ENSBGRP00000023084 | 96.8593 | 94.9749 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 97.3684 | 95.614 | chinchilla_lanigera | ENSCLAP00000002859 | 97.6131 | 95.8543 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 96.4912 | 94.4862 | cervus_hanglu_yarkandensis | ENSCHYP00000032246 | 94.0171 | 92.0635 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 96.4912 | 94.8622 | catagonus_wagneri | ENSCWAP00000028762 | 96.7337 | 95.1005 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 96.8672 | 94.6115 | ictidomys_tridecemlineatus | ENSSTOP00000026378 | 96.264 | 94.0224 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 97.4937 | 96.1153 | otolemur_garnettii | ENSOGAP00000013047 | 97.4937 | 96.1153 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 98.7469 | 98.4962 | cebus_imitator | ENSCCAP00000013880 | 97.0443 | 96.798 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 97.7444 | 95.8647 | urocitellus_parryii | ENSUPAP00010022971 | 97.99 | 96.1055 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 96.4912 | 94.6115 | moschus_moschiferus | ENSMMSP00000018815 | 96.6123 | 94.7302 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 99.4987 | 99.4987 | rhinopithecus_roxellana | ENSRROP00000002581 | 99.4987 | 99.4987 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 96.8672 | 94.2356 | mus_pahari | MGP_PahariEiJ_P0031343 | 97.1105 | 94.4724 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 98.6216 | 97.4937 | propithecus_coquereli | ENSPCOP00000021655 | 97.2806 | 96.1681 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 33.208 | 17.6692 | drosophila_melanogaster | FBpp0078586 | 24.836 | 13.2146 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 22.6817 | 14.6617 | eptatretus_burgeri | ENSEBUP00000023000 | 44.9132 | 29.0323 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 78.6967 | 67.7945 | callorhinchus_milii | ENSCMIP00000023791 | 76.399 | 65.8151 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 81.8296 | 73.0576 | latimeria_chalumnae | ENSLACP00000015962 | 83.3972 | 74.4572 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 77.9449 | 67.4185 | lepisosteus_oculatus | ENSLOCP00000015207 | 74.6699 | 64.5858 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 67.6692 | 57.3935 | danio_rerio | ENSDARP00000088481 | 62.8638 | 53.3178 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 70.802 | 59.7744 | carassius_auratus | ENSCARP00000121498 | 49.6049 | 41.8788 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 70.1754 | 59.1479 | carassius_auratus | ENSCARP00000049293 | 47.2973 | 39.8649 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 73.3083 | 60.6516 | astyanax_mexicanus | ENSAMXP00000031035 | 66.7047 | 55.1881 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 71.6792 | 58.396 | ictalurus_punctatus | ENSIPUP00000030071 | 49.4382 | 40.2766 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 71.4286 | 61.4035 | esox_lucius | ENSELUP00000017291 | 61.2245 | 52.6316 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 70.1754 | 57.8947 | electrophorus_electricus | ENSEEEP00000007442 | 64.2939 | 53.0425 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 71.9298 | 61.2782 | oncorhynchus_kisutch | ENSOKIP00005055175 | 60.9342 | 51.9108 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 71.6792 | 61.9048 | oncorhynchus_kisutch | ENSOKIP00005007700 | 45.2174 | 39.0514 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 72.0551 | 61.2782 | oncorhynchus_mykiss | ENSOMYP00000122673 | 45.24 | 38.4736 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 71.8045 | 61.9048 | oncorhynchus_mykiss | ENSOMYP00000101759 | 44.4531 | 38.3243 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 71.9298 | 62.406 | salmo_salar | ENSSSAP00000161873 | 45.3755 | 39.3676 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 41.3534 | 35.3383 | salmo_salar | ENSSSAP00000087329 | 35.5603 | 30.3879 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 72.0551 | 62.406 | salmo_trutta | ENSSTUP00000092688 | 45.4186 | 39.3365 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 72.4311 | 61.5288 | salmo_trutta | ENSSTUP00000004496 | 60.7143 | 51.5756 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 71.3033 | 60.401 | fundulus_heteroclitus | ENSFHEP00000034300 | 44.8032 | 37.9528 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 71.5539 | 60.2757 | poecilia_reticulata | ENSPREP00000028362 | 44.4704 | 37.4611 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 71.6792 | 60.401 | xiphophorus_maculatus | ENSXMAP00000007781 | 45.1104 | 38.0126 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 71.5539 | 61.1529 | oryzias_latipes | ENSORLP00000006028 | 45.0315 | 38.4858 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 70.3008 | 59.6491 | cyclopterus_lumpus | ENSCLMP00005022101 | 60.5178 | 51.3484 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 72.9323 | 61.4035 | oreochromis_niloticus | ENSONIP00000027136 | 45.3271 | 38.162 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 72.807 | 61.7794 | haplochromis_burtoni | ENSHBUP00000003949 | 45.2845 | 38.4256 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 71.5539 | 60.6516 | astatotilapia_calliptera | ENSACLP00000001514 | 44.7843 | 37.9608 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 73.0576 | 62.1554 | sparus_aurata | ENSSAUP00010067091 | 45.7614 | 38.9325 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 73.8095 | 62.9073 | lates_calcarifer | ENSLCAP00010036265 | 61.9348 | 52.7865 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 84.4612 | 76.5664 | xenopus_tropicalis | ENSXETP00000069597 | 85.7506 | 77.7354 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 30.5764 | 28.4461 | chrysemys_picta_bellii | ENSCPBP00000040697 | 75.7764 | 70.4969 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 92.7318 | 87.8446 | sphenodon_punctatus | ENSSPUP00000017594 | 93.0818 | 88.1761 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 90.4762 | 85.4637 | crocodylus_porosus | ENSCPRP00005006856 | 92.9215 | 87.7735 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 25.9398 | 22.6817 | laticauda_laticaudata | ENSLLTP00000021739 | 68.0921 | 59.5395 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 46.2406 | 39.3484 | notechis_scutatus | ENSNSUP00000014341 | 74.5455 | 63.4343 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 21.3033 | 18.0451 | pseudonaja_textilis | ENSPTXP00000020768 | 74.8899 | 63.4361 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 54.5113 | 46.6165 | pseudonaja_textilis | ENSPTXP00000020766 | 75.784 | 64.8084 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 92.3559 | 87.3434 | anas_platyrhynchos_platyrhynchos | ENSAPLP00000031219 | 88.3693 | 83.5731 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 91.9799 | 86.8421 | gallus_gallus | ENSGALP00010013622 | 89.2944 | 84.3066 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 91.4787 | 86.7168 | meleagris_gallopavo | ENSMGAP00000013155 | 90.5707 | 85.8561 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 32.5815 | 29.0727 | serinus_canaria | ENSSCAP00000005948 | 88.1356 | 78.6441 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 92.4812 | 87.7193 | parus_major | ENSPMJP00000021974 | 92.8302 | 88.0503 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 89.0977 | 83.208 | anolis_carolinensis | ENSACAP00000000888 | 89.2095 | 83.3124 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 67.1679 | 57.7694 | neolamprologus_brichardi | ENSNBRP00000030776 | 70.4336 | 60.5782 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 53.7594 | 45.614 | naja_naja | ENSNNAP00000022023 | 76.3345 | 64.7687 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 90.4762 | 83.8346 | podarcis_muralis | ENSPMRP00000019331 | 90.5897 | 83.9398 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 64.1604 | 60.0251 | geospiza_fortis | ENSGFOP00000001981 | 90.1408 | 84.331 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 31.0777 | 28.4461 | geospiza_fortis | ENSGFOP00000001994 | 90.1818 | 82.5455 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 91.1028 | 85.7143 | taeniopygia_guttata | ENSTGUP00000035013 | 90.0867 | 84.7584 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 74.0602 | 64.9123 | erpetoichthys_calabaricus | ENSECRP00000004191 | 72.3378 | 63.4027 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 72.1805 | 60.6516 | kryptolebias_marmoratus | ENSKMAP00000007182 | 61.2766 | 51.4894 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 89.8496 | 83.0827 | salvator_merianae | ENSSMRP00000001855 | 90.3023 | 83.5013 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 50 | 41.1028 | oryzias_javanicus | ENSOJAP00000026905 | 38.4023 | 31.5688 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 65.1629 | 56.391 | amphilophus_citrinellus | ENSACIP00000012507 | 69.5187 | 60.1604 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 72.807 | 62.406 | dicentrarchus_labrax | ENSDLAP00005051742 | 44.7957 | 38.3963 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 68.5464 | 59.1479 | amphiprion_percula | ENSAPEP00000031817 | 67.0343 | 57.8431 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 71.4286 | 60.7769 | oryzias_sinensis | ENSOSIP00000036190 | 45.0593 | 38.3399 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 92.8571 | 88.0952 | pelodiscus_sinensis | ENSPSIP00000013344 | 93.2076 | 88.4277 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 91.8546 | 86.5915 | gopherus_evgoodei | ENSGEVP00005025215 | 92.3174 | 87.0277 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 90.1003 | 84.8371 | chelonoidis_abingdonii | ENSCABP00000031679 | 91.128 | 85.8048 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 67.0426 | 57.7694 | seriola_dumerili | ENSSDUP00000010144 | 69.3904 | 59.7925 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 74.0602 | 62.6566 | cottoperca_gobio | ENSCGOP00000025817 | 46.1719 | 39.0625 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 93.985 | 89.2231 | struthio_camelus_australis | ENSSCUP00000008929 | 94.3396 | 89.5597 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 90.1003 | 84.8371 | anser_brachyrhynchus | ENSABRP00000016867 | 89.4279 | 84.204 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 33.4586 | 27.3183 | gasterosteus_aculeatus | ENSGACP00000025838 | 54.9383 | 44.856 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 71.5539 | 61.4035 | cynoglossus_semilaevis | ENSCSEP00000010745 | 45.2456 | 38.8273 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 70.4261 | 60.0251 | poecilia_formosa | ENSPFOP00000001373 | 60.3652 | 51.4501 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 73.5589 | 62.5313 | myripristis_murdjan | ENSMMDP00005031023 | 61.2735 | 52.0877 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 73.183 | 62.1554 | sander_lucioperca | ENSSLUP00000053923 | 45.9119 | 38.9937 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 92.6065 | 87.9699 | strigops_habroptila | ENSSHBP00005020872 | 92.956 | 88.3019 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 73.183 | 62.6566 | amphiprion_ocellaris | ENSAOCP00000003446 | 45.4121 | 38.8802 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 92.8571 | 87.9699 | terrapene_carolina_triunguis | ENSTMTP00000003246 | 93.2076 | 88.3019 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 70.802 | 60.7769 | hucho_hucho | ENSHHUP00000016304 | 56.1073 | 48.1629 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 69.1729 | 58.396 | hucho_hucho | ENSHHUP00000081847 | 43.5331 | 36.7508 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 73.0576 | 61.5288 | betta_splendens | ENSBSLP00000036925 | 57.7228 | 48.6139 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 73.6842 | 61.5288 | pygocentrus_nattereri | ENSPNAP00000003930 | 66.7423 | 55.7321 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 73.5589 | 62.406 | anabas_testudineus | ENSATEP00000024102 | 61.6597 | 52.3109 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 70.802 | 60.7769 | seriola_lalandi_dorsalis | ENSSLDP00000008842 | 62.362 | 53.532 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 70.3008 | 59.6491 | labrus_bergylta | ENSLBEP00000025662 | 56.724 | 48.1294 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 72.807 | 61.7794 | pundamilia_nyererei | ENSPNYP00000009022 | 45.2845 | 38.4256 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 72.4311 | 62.5313 | mastacembelus_armatus | ENSMAMP00000008130 | 60.9705 | 52.6371 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 72.4311 | 61.4035 | stegastes_partitus | ENSSPAP00000031370 | 45.3333 | 38.4314 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 21.6792 | 18.6717 | oncorhynchus_tshawytscha | ENSOTSP00005104594 | 65.0376 | 56.015 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 71.9298 | 61.1529 | oncorhynchus_tshawytscha | ENSOTSP00005102904 | 45.1258 | 38.3648 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 46.8672 | 39.8496 | oncorhynchus_tshawytscha | ENSOTSP00005104590 | 41.0989 | 34.9451 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 73.4336 | 62.6566 | acanthochromis_polyacanthus | ENSAPOP00000031414 | 45.5676 | 38.8802 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 93.4837 | 88.5965 | aquila_chrysaetos_chrysaetos | ENSACCP00020000334 | 93.8365 | 88.9308 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 71.5539 | 59.7744 | nothobranchius_furzeri | ENSNFUP00015050020 | 45.1027 | 37.6777 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 45.99 | 37.594 | cyprinodon_variegatus | ENSCVAP00000029047 | 41.1435 | 33.6323 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 70.6767 | 60.2757 | denticeps_clupeoides | ENSDCDP00000014274 | 62.9464 | 53.683 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 73.183 | 62.0301 | takifugu_rubripes | ENSTRUP00000017751 | 44.7167 | 37.902 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 92.6065 | 87.3434 | ficedula_albicollis | ENSFALP00000021417 | 92.956 | 87.673 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 67.9198 | 58.1454 | hippocampus_comes | ENSHCOP00000011947 | 59.235 | 50.7104 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 71.3033 | 60.6516 | cyprinus_carpio_carpio | ENSCCRP00000166469 | 49.9561 | 42.4934 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 70.9273 | 59.8997 | cyprinus_carpio_carpio | ENSCCRP00000141699 | 48.9619 | 41.3495 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 91.3534 | 86.5915 | coturnix_japonica | ENSCJPP00005023131 | 90.6716 | 85.9453 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 63.5338 | 54.386 | poecilia_latipinna | ENSPLAP00000001710 | 64.9168 | 55.5698 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 70.9273 | 59.8997 | sinocyclocheilus_grahami | ENSSGRP00000056358 | 64.3914 | 54.38 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 70.6767 | 59.2732 | sinocyclocheilus_grahami | ENSSGRP00000049177 | 47.6351 | 39.9493 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 72.6817 | 61.6541 | maylandia_zebra | ENSMZEP00005002173 | 45.2065 | 38.3476 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 19.5489 | 17.6692 | tetraodon_nigroviridis | ENSTNIP00000004614 | 93.9759 | 84.9398 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 31.9549 | 26.3158 | tetraodon_nigroviridis | ENSTNIP00000014268 | 57.5621 | 47.4041 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 67.2932 | 55.2632 | paramormyrops_kingsleyae | ENSPKIP00000017908 | 47.3963 | 38.9232 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 66.2907 | 54.1353 | paramormyrops_kingsleyae | ENSPKIP00000016034 | 47.2744 | 38.6059 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 68.2957 | 55.8897 | scleropages_formosus | ENSSFOP00015002420 | 56.6528 | 46.3617 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 62.406 | 50.1253 | scleropages_formosus | ENSSFOP00015028870 | 65.1832 | 52.356 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 84.3358 | 76.0652 | leptobrachium_leishanense | ENSLLEP00000005123 | 85.7325 | 77.3248 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 72.6817 | 61.5288 | scophthalmus_maximus | ENSSMAP00000018886 | 45.4189 | 38.4495 |
| ENSG00000109819 | homo_sapiens | ENSP00000264867 | 70.3008 | 59.7744 | oryzias_melastigma | ENSOMEP00000005014 | 60.3875 | 51.3455 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0000785 | part_of chromatin | 1 | IC | Homo_sapiens(9606) | Component |
| GO:0001659 | involved_in temperature homeostasis | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0001678 | involved_in intracellular glucose homeostasis | 1 | NAS | Homo_sapiens(9606) | Process |
| GO:0002021 | involved_in response to dietary excess | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0003677 | enables DNA binding | 2 | ISS,TAS | Homo_sapiens(9606) | Function |
| GO:0003712 | enables transcription coregulator activity | 2 | ISS,TAS | Homo_sapiens(9606) | Function |
| GO:0003713 | enables transcription coactivator activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0003723 | enables RNA binding | 1 | TAS | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005634 | is_active_in nucleus | 5 | IBA,IDA,ISS,TAS | Homo_sapiens(9606) | Component |
| GO:0005654 | located_in nucleoplasm | 2 | IDA,TAS | Homo_sapiens(9606) | Component |
| GO:0005829 | located_in cytosol | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0006094 | involved_in gluconeogenesis | 2 | IDA,NAS | Homo_sapiens(9606) | Process |
| GO:0006355 | acts_upstream_of_or_within regulation of DNA-templated transcription | 2 | IDA | Homo_sapiens(9606) | Process |
| GO:0006367 | involved_in transcription initiation at RNA polymerase II promoter | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0006397 | involved_in mRNA processing | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0007005 | involved_in mitochondrion organization | 1 | NAS | Homo_sapiens(9606) | Process |
| GO:0007586 | involved_in digestion | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0008134 | enables transcription factor binding | 1 | IBA | Homo_sapiens(9606) | Function |
| GO:0008380 | involved_in RNA splicing | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0010628 | involved_in positive regulation of gene expression | 2 | IDA,IMP | Homo_sapiens(9606) | Process |
| GO:0010822 | involved_in positive regulation of mitochondrion organization | 2 | IMP,ISS | Homo_sapiens(9606) | Process |
| GO:0014850 | involved_in response to muscle activity | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0016605 | located_in PML body | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0016922 | enables nuclear receptor binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0019395 | involved_in fatty acid oxidation | 1 | NAS | Homo_sapiens(9606) | Process |
| GO:0022904 | involved_in respiratory electron transport chain | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0030374 | enables nuclear receptor coactivator activity | 2 | IBA,IDA | Homo_sapiens(9606) | Function |
| GO:0031490 | enables chromatin DNA binding | 1 | ISS | Homo_sapiens(9606) | Function |
| GO:0031625 | enables ubiquitin protein ligase binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0032922 | involved_in circadian regulation of gene expression | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0034599 | involved_in cellular response to oxidative stress | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0035066 | involved_in positive regulation of histone acetylation | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0042594 | involved_in response to starvation | 1 | NAS | Homo_sapiens(9606) | Process |
| GO:0042752 | involved_in regulation of circadian rhythm | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0043524 | involved_in negative regulation of neuron apoptotic process | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0043565 | enables sequence-specific DNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0045333 | involved_in cellular respiration | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0045722 | involved_in positive regulation of gluconeogenesis | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0045893 | involved_in positive regulation of DNA-templated transcription | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0045944 | involved_in positive regulation of transcription by RNA polymerase II | 3 | IBA,IDA,ISS | Homo_sapiens(9606) | Process |
| GO:0046321 | involved_in positive regulation of fatty acid oxidation | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0048662 | acts_upstream_of negative regulation of smooth muscle cell proliferation | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0050821 | involved_in protein stabilization | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0050873 | involved_in brown fat cell differentiation | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0051091 | involved_in positive regulation of DNA-binding transcription factor activity | 2 | IBA,IDA | Homo_sapiens(9606) | Process |
| GO:0051402 | involved_in neuron apoptotic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0060612 | involved_in adipose tissue development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0061629 | enables RNA polymerase II-specific DNA-binding transcription factor binding | 1 | ISS | Homo_sapiens(9606) | Function |
| GO:0065003 | involved_in protein-containing complex assembly | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0097009 | involved_in energy homeostasis | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0106222 | enables lncRNA binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0120162 | involved_in positive regulation of cold-induced thermogenesis | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0140297 | enables DNA-binding transcription factor binding | 1 | TAS | Homo_sapiens(9606) | Function |
| GO:1901857 | involved_in positive regulation of cellular respiration | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:1901860 | involved_in positive regulation of mitochondrial DNA metabolic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:1901863 | involved_in positive regulation of muscle tissue development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:2000277 | involved_in positive regulation of oxidative phosphorylation uncoupler activity | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:2001171 | involved_in positive regulation of ATP biosynthetic process | 1 | ISS | Homo_sapiens(9606) | Process |