RBP Type
Canonical_RBPs
Diseases
N.A.
Drug
Main interacting RNAs
N.A.
Moonlighting functions
N.A.
Localizations
N.A.
BulkPerturb-seq
Ensembl ID ENSG00000269858 Gene ID
112398 Accession
14660
Symbol
EGLN2
Alias
EIT6;PHD1;EIT-6;HPH-1;HPH-3;HIFPH1;HIF-PH1
Full Name
egl-9 family hypoxia inducible factor 2
Status
Confidence
Length
9439 bases
Strand
Plus strand
Position
19 : 40798996 - 40808434
RNA binding domain
2OG-FeII_Oxy
Summary
The hypoxia inducible factor (HIF) is a transcriptional complex that is involved in oxygen homeostasis. At normal oxygen levels, the alpha subunit of HIF is targeted for degration by prolyl hydroxylation. This gene encodes an enzyme responsible for this post-translational modification. Alternative splicing results in multiple transcript variants. Read-through transcription also exists between this gene and the upstream RAB4B (RAB4B, member RAS oncogene family) gene. [provided by RefSeq, Feb 2011]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
ENSP00000472414
2OG-FeII_Oxy
PF03171 0.00019
1
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
Name
Transcript ID
bp
Protein
Translation ID
EGLN2-215
ENST00000598654
590
101aa
ENSP00000471568
EGLN2-201
ENST00000303961
2100
407aa
ENSP00000307080
EGLN2-208
ENST00000593972
935
139aa
ENSP00000471546
EGLN2-202
ENST00000406058
2150
407aa
ENSP00000385253
EGLN2-207
ENST00000593726
2821
407aa
ENSP00000469686
EGLN2-210
ENST00000594380
597
87aa
ENSP00000470968
EGLN2-203
ENST00000593397
572
72aa
ENSP00000472146
EGLN2-217
ENST00000601733
584
94aa
ENSP00000469323
EGLN2-206
ENST00000593525
709
77aa
ENSP00000469272
EGLN2-213
ENST00000596517
574
67aa
ENSP00000472870
EGLN2-214
ENST00000597746
459
128aa
ENSP00000472991
EGLN2-209
ENST00000594140
991
125aa
ENSP00000472414
EGLN2-216
ENST00000599579
3894
No protein
-
EGLN2-204
ENST00000593445
3034
No protein
-
EGLN2-205
ENST00000593477
1391
No protein
-
EGLN2-218
ENST00000602166
684
No protein
-
EGLN2-212
ENST00000595621
758
96aa
ENSP00000471944
EGLN2-211
ENST00000595051
477
51aa
ENSP00000471740
Pathway ID
Pathway Name
Source
ensgID
Trait
pValue
Pubmed ID
65186 Bronchodilator Agents 3E-6 26634245 65187 Forced Expiratory Volume 3E-6 26634245 68922 Bronchodilator Agents 4E-6 26634245 68923 Forced Expiratory Volume 4E-6 26634245 74945 Bronchodilator Agents 4E-6 26634245 74946 Forced Expiratory Volume 4E-6 26634245 76846 Bronchodilator Agents 4E-6 26634245 76847 Forced Expiratory Volume 4E-6 26634245 96791 Bronchodilator Agents 5E-6 26634245 96792 Forced Expiratory Volume 5E-6 26634245 103180 Smoking 1E-8 20418890
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
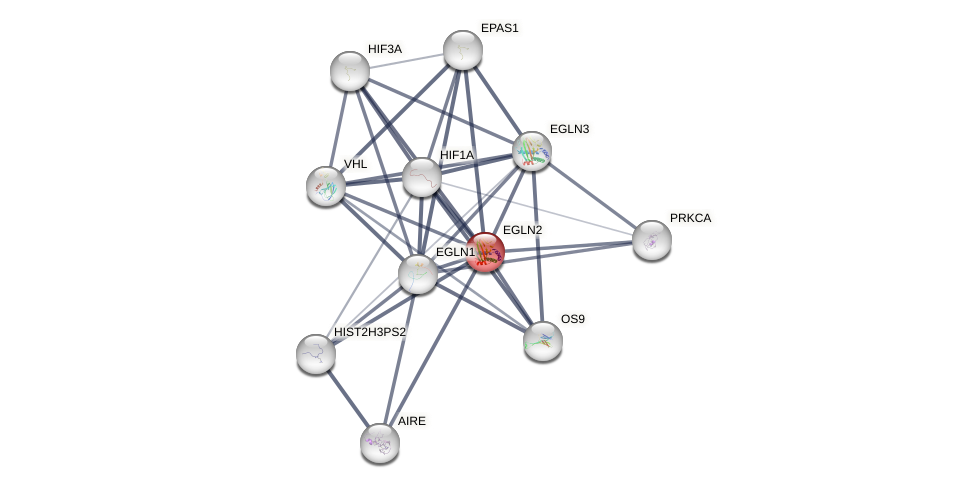
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0001558 involved_in regulation of cell growth 1 NAS Homo_sapiens(9606) Process GO:0001666 involved_in response to hypoxia 1 IDA Homo_sapiens(9606) Process GO:0005515 enables protein binding 1 IPI Homo_sapiens(9606) Function GO:0005634 is_active_in nucleus 2 IBA,IDA Homo_sapiens(9606) Component GO:0005654 located_in nucleoplasm 2 IDA,TAS Homo_sapiens(9606) Component GO:0005737 is_active_in cytoplasm 1 IBA Homo_sapiens(9606) Component GO:0008198 enables ferrous iron binding 3 IBA,IDA,NAS Homo_sapiens(9606) Function GO:0016706 enables 2-oxoglutarate-dependent dioxygenase activity 1 IDA Homo_sapiens(9606) Function GO:0018401 involved_in peptidyl-proline hydroxylation to 4-hydroxy-L-proline 2 IBA,IDA Homo_sapiens(9606) Process GO:0019826 enables oxygen sensor activity 1 IDA Homo_sapiens(9606) Function GO:0030520 involved_in intracellular estrogen receptor signaling pathway 1 NAS Homo_sapiens(9606) Process GO:0031418 enables L-ascorbic acid binding 1 IEA Homo_sapiens(9606) Function GO:0031545 enables peptidyl-proline 4-dioxygenase activity 2 IBA,IDA Homo_sapiens(9606) Function GO:0043523 involved_in regulation of neuron apoptotic process 1 IMP Homo_sapiens(9606) Process GO:0045454 involved_in cell redox homeostasis 1 IDA Homo_sapiens(9606) Process GO:0045732 involved_in positive regulation of protein catabolic process 1 IDA Homo_sapiens(9606) Process GO:0071456 involved_in cellular response to hypoxia 1 IBA Homo_sapiens(9606) Process
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.