| GO:0000139 | located_in Golgi membrane | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0001774 | involved_in microglial cell activation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0001932 | involved_in regulation of protein phosphorylation | 1 | IEA | Homo_sapiens(9606) | Process |
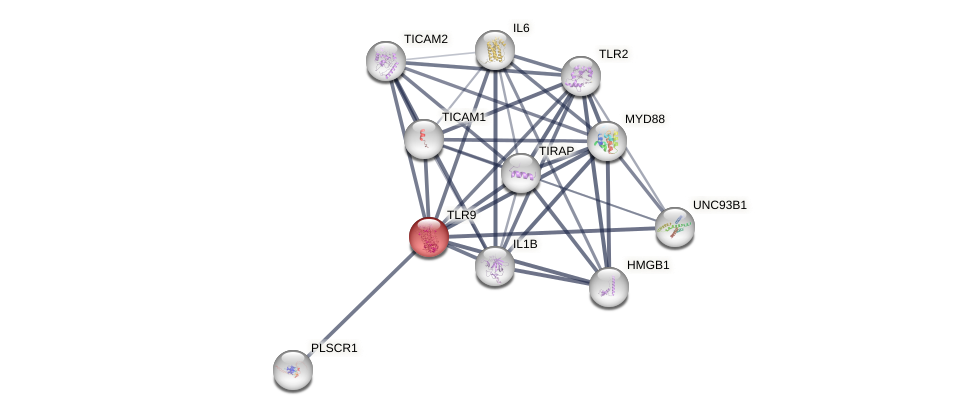
| GO:0002224 | involved_in toll-like receptor signaling pathway | 2 | IBA,IDA | Homo_sapiens(9606) | Process |
| GO:0002639 | involved_in positive regulation of immunoglobulin production | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0002730 | involved_in regulation of dendritic cell cytokine production | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0002755 | involved_in MyD88-dependent toll-like receptor signaling pathway | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0005149 | enables interleukin-1 receptor binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005576 | located_in extracellular region | 1 | NAS | Homo_sapiens(9606) | Component |
| GO:0005737 | located_in cytoplasm | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005764 | located_in lysosome | 1 | ISS | Homo_sapiens(9606) | Component |
| GO:0005768 | located_in endosome | 1 | ISS | Homo_sapiens(9606) | Component |
| GO:0005783 | is_active_in endoplasmic reticulum | 2 | IDA,ISS | Homo_sapiens(9606) | Component |
| GO:0005789 | located_in endoplasmic reticulum membrane | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0005886 | is_active_in plasma membrane | 2 | IBA,IDA | Homo_sapiens(9606) | Component |
| GO:0007249 | involved_in I-kappaB kinase/NF-kappaB signaling | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0008584 | involved_in male gonad development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0010008 | located_in endosome membrane | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0010508 | involved_in positive regulation of autophagy | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0010628 | involved_in positive regulation of gene expression | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0016323 | located_in basolateral plasma membrane | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0016324 | located_in apical plasma membrane | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0030277 | involved_in maintenance of gastrointestinal epithelium | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0030890 | involved_in positive regulation of B cell proliferation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0032009 | located_in early phagosome | 1 | ISS | Homo_sapiens(9606) | Component |
| GO:0032490 | involved_in detection of molecule of bacterial origin | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0032722 | involved_in positive regulation of chemokine production | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0032725 | involved_in positive regulation of granulocyte macrophage colony-stimulating factor production | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0032727 | involved_in positive regulation of interferon-alpha production | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0032728 | involved_in positive regulation of interferon-beta production | 2 | IDA,ISS | Homo_sapiens(9606) | Process |
| GO:0032729 | involved_in positive regulation of type II interferon production | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0032733 | involved_in positive regulation of interleukin-10 production | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0032735 | involved_in positive regulation of interleukin-12 production | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0032741 | involved_in positive regulation of interleukin-18 production | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0032755 | involved_in positive regulation of interleukin-6 production | 2 | IBA,IDA | Homo_sapiens(9606) | Process |
| GO:0032757 | acts_upstream_of positive regulation of interleukin-8 production | 2 | IDA | Homo_sapiens(9606) | Process |
| GO:0032760 | involved_in positive regulation of tumor necrosis factor production | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0034162 | involved_in toll-like receptor 9 signaling pathway | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0034163 | involved_in regulation of toll-like receptor 9 signaling pathway | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0034165 | involved_in positive regulation of toll-like receptor 9 signaling pathway | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0035197 | enables siRNA binding | 1 | IMP | Homo_sapiens(9606) | Function |
| GO:0036019 | located_in endolysosome | 1 | ISS | Homo_sapiens(9606) | Component |
| GO:0036020 | located_in endolysosome membrane | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0038187 | enables pattern recognition receptor activity | 2 | IBA,IDA | Homo_sapiens(9606) | Function |
| GO:0042742 | involved_in defense response to bacterium | 1 | NAS | Homo_sapiens(9606) | Process |
| GO:0042803 | enables protein homodimerization activity | 1 | ISS | Homo_sapiens(9606) | Function |
| GO:0043123 | involved_in positive regulation of I-kappaB kinase/NF-kappaB signaling | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0043410 | involved_in positive regulation of MAPK cascade | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0045087 | involved_in innate immune response | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0045322 | enables unmethylated CpG binding | 1 | ISS | Homo_sapiens(9606) | Function |
| GO:0045577 | involved_in regulation of B cell differentiation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0045944 | involved_in positive regulation of transcription by RNA polymerase II | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0046330 | involved_in positive regulation of JNK cascade | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0050729 | involved_in positive regulation of inflammatory response | 1 | IC | Homo_sapiens(9606) | Process |
| GO:0050829 | involved_in defense response to Gram-negative bacterium | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0050871 | involved_in positive regulation of B cell activation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0051092 | involved_in positive regulation of NF-kappaB transcription factor activity | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0051607 | involved_in defense response to virus | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0070373 | involved_in negative regulation of ERK1 and ERK2 cascade | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0071222 | involved_in cellular response to lipopolysaccharide | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0071248 | involved_in cellular response to metal ion | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:1901224 | involved_in positive regulation of NIK/NF-kappaB signaling | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:1901895 | involved_in negative regulation of ATPase-coupled calcium transmembrane transporter activity | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:1902350 | involved_in cellular response to chloroquine | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:1905300 | involved_in positive regulation of intestinal epithelial cell development | 1 | ISS | Homo_sapiens(9606) | Process |