RBP Type
Canonical_RBPs
Diseases
Drug
Main interacting RNAs
N.A.
Moonlighting functions
N.A.
Localizations
N.A.
BulkPerturb-seq
Ensembl ID ENSG00000198838 Gene ID
6263 Accession
10485
Symbol
RYR3
Alias
RYR-3;CMYP20
Full Name
ryanodine receptor 3
Status
Confidence
Length
555160 bases
Strand
Plus strand
Position
15 : 33310962 - 33866121
RNA binding domain
SPRY
Summary
The protein encoded by this gene is a ryanodine receptor, which functions to release calcium from intracellular storage for use in many cellular processes. For example, the encoded protein is involved in skeletal muscle contraction by releasing calcium from the sarcoplasmic reticulum followed by depolarization of T-tubules. Two transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Sep 2011]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
ENSP00000489529
SPRY
PF00622 3.2e-62
1
ENSP00000489529
SPRY
PF00622 3.2e-62
2
ENSP00000489529
SPRY
PF00622 3.2e-62
3
ENSP00000399610
SPRY
PF00622 3.2e-62
1
ENSP00000399610
SPRY
PF00622 3.2e-62
2
ENSP00000399610
SPRY
PF00622 3.2e-62
3
ENSP00000373884
SPRY
PF00622 3.3e-62
1
ENSP00000373884
SPRY
PF00622 3.3e-62
2
ENSP00000373884
SPRY
PF00622 3.3e-62
3
ENSP00000489262
SPRY
PF00622 3.3e-62
1
ENSP00000489262
SPRY
PF00622 3.3e-62
2
ENSP00000489262
SPRY
PF00622 3.3e-62
3
ENSP00000483166
SPRY
PF00622 3.3e-62
1
ENSP00000483166
SPRY
PF00622 3.3e-62
2
ENSP00000483166
SPRY
PF00622 3.3e-62
3
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
26309413 A polymorphism at the microRNA binding site in the 3' untranslated region of RYR3 is associated with outcome in hepatocellular carcinoma.
Chenxing Peng
2015-01-01
OncoTargets and therapy
Name
Transcript ID
bp
Protein
Translation ID
RYR3-212
ENST00000634891
15568
4870aa
ENSP00000489262
RYR3-202
ENST00000415757
15557
4865aa
ENSP00000399610
RYR3-201
ENST00000389232
15552
4869aa
ENSP00000373884
RYR3-209
ENST00000634418
15078
4859aa
ENSP00000489529
RYR3-211
ENST00000634750
137
46aa
ENSP00000489294
RYR3-216
ENST00000635875
100
No protein
-
RYR3-222
ENST00000636753
100
No protein
-
RYR3-220
ENST00000636583
100
No protein
-
RYR3-214
ENST00000635790
8528
1101aa
ENSP00000489672
RYR3-217
ENST00000636417
100
No protein
-
RYR3-226
ENST00000637201
100
No protein
-
RYR3-234
ENST00000638085
143
No protein
-
RYR3-232
ENST00000638038
100
No protein
-
RYR3-203
ENST00000557931
161
No protein
-
RYR3-204
ENST00000558060
451
No protein
-
RYR3-235
ENST00000638145
100
No protein
-
RYR3-213
ENST00000635749
3714
No protein
-
RYR3-215
ENST00000635842
100
33aa
ENSP00000490438
RYR3-231
ENST00000638032
100
34aa
ENSP00000489937
RYR3-229
ENST00000637948
3026
284aa
ENSP00000489832
RYR3-223
ENST00000636845
2973
454aa
ENSP00000490033
RYR3-210
ENST00000634730
2743
615aa
ENSP00000489346
RYR3-230
ENST00000637984
100
No protein
-
RYR3-228
ENST00000637615
100
No protein
-
RYR3-233
ENST00000638052
1874
No protein
-
RYR3-221
ENST00000636656
100
No protein
-
RYR3-219
ENST00000636568
2257
468aa
ENSP00000490632
RYR3-227
ENST00000637522
137
No protein
-
RYR3-206
ENST00000559917
702
No protein
-
RYR3-205
ENST00000559333
374
No protein
-
RYR3-225
ENST00000637072
3156
No protein
-
RYR3-207
ENST00000560791
581
No protein
-
RYR3-218
ENST00000636497
100
No protein
-
RYR3-224
ENST00000636878
149
No protein
-
RYR3-208
ENST00000622037
15564
4873aa
ENSP00000483166
Pathway ID
Pathway Name
Source
ensgID
Trait
pValue
Pubmed ID
7825 Creatinine 6.38799512153732E-5 17903292 7870 Creatinine 6.41503010230249E-5 17903292 10190 Exercise Test 5.19399492631152E-7 17903301 15664 Stroke 9.5485844E-004 - 15749 Stroke 2.1562131E-004 - 18300 Fetal Hemoglobin 5.142e-007 - 19536 Blood Pressure 8.041e-005 - 20538 Bilirubin 9.681e-005 - 20540 Bilirubin 3.562e-005 - 20541 Bilirubin 4.662e-005 - 20596 Bilirubin 4.662e-005 - 20636 Bilirubin 4.662e-005 - 27174 Blood Pressure 6.9850000E-005 - 28164 Blood Glucose 1.2810000E-005 - 28819 monocyte chemoattractant protein 1 (66-77) 5.2190000E-005 - 29344 Cholesterol, LDL 7.3990000E-005 - 30804 Erythrocyte Indices 5.3030000E-005 - 33070 Ankle Brachial Index 8.4875008E-005 - 33830 Glomerular Filtration Rate 3.6067547E-005 - 34561 Platelet Function Tests 7.0600000E-006 - 34587 Platelet Function Tests 1.4493300E-005 - 36445 Platelet Function Tests 3.8654100E-005 - 37689 Platelet Function Tests 2.2120700E-005 - 44632 Platelet Function Tests 1.6799400E-005 - 45977 Platelet Function Tests 4.0005362E-006 - 46192 Platelet Function Tests 1.5443800E-005 - 49279 Child Development Disorders, Pervasive 7.4618000E-005 - 50335 Autistic Disorder 2.4901000E-005 - 50339 Autistic Disorder 3.2432000E-005 - 71004 Longevity 6E-6 27015805 71791 Trans Fatty Acids 2E-6 25646338 76397 HIV-1 3E-8 20009918 76398 Vascular Calcification 3E-8 20009918 80591 Trans Fatty Acids 1E-6 25646338 83323 Diabetic Nephropathies 9E-6 26305897 84948 Trans Fatty Acids 1E-6 25646338 96429 Trans Fatty Acids 1E-6 25646338
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
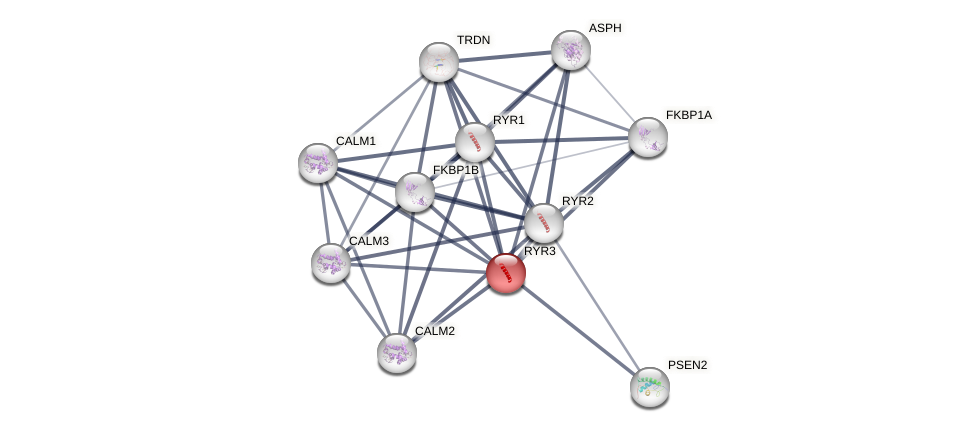
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0005219 enables ryanodine-sensitive calcium-release channel activity 3 IBA,ISS,TAS Homo_sapiens(9606) Function GO:0005509 enables calcium ion binding 1 IEA Homo_sapiens(9606) Function GO:0005516 enables calmodulin binding 1 IEA Homo_sapiens(9606) Function GO:0005790 is_active_in smooth endoplasmic reticulum 1 IBA Homo_sapiens(9606) Component GO:0006816 involved_in calcium ion transport 1 IMP Homo_sapiens(9606) Process GO:0006874 involved_in intracellular calcium ion homeostasis 1 IEA Homo_sapiens(9606) Process GO:0006941 involved_in striated muscle contraction 1 IBA Homo_sapiens(9606) Process GO:0014808 involved_in release of sequestered calcium ion into cytosol by sarcoplasmic reticulum 1 IBA Homo_sapiens(9606) Process GO:0015278 enables calcium-release channel activity 1 IMP Homo_sapiens(9606) Function GO:0016020 located_in membrane 1 ISS Homo_sapiens(9606) Component GO:0016529 located_in sarcoplasmic reticulum 1 ISS Homo_sapiens(9606) Component GO:0030018 is_active_in Z disc 1 IBA Homo_sapiens(9606) Component GO:0033017 is_active_in sarcoplasmic reticulum membrane 3 IBA,ISS,TAS Homo_sapiens(9606) Component GO:0034704 part_of calcium channel complex 1 IBA Homo_sapiens(9606) Component GO:0042383 is_active_in sarcolemma 1 IBA Homo_sapiens(9606) Component GO:0048763 enables calcium-induced calcium release activity 1 IBA Homo_sapiens(9606) Function GO:0051289 involved_in protein homotetramerization 1 ISS Homo_sapiens(9606) Process GO:0070588 involved_in calcium ion transmembrane transport 1 ISS Homo_sapiens(9606) Process GO:0071277 involved_in cellular response to calcium ion 1 ISS Homo_sapiens(9606) Process GO:0071286 involved_in cellular response to magnesium ion 1 ISS Homo_sapiens(9606) Process GO:0071313 involved_in cellular response to caffeine 1 ISS Homo_sapiens(9606) Process GO:0071318 involved_in cellular response to ATP 1 ISS Homo_sapiens(9606) Process
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.