RBP Type
Canonical_RBPs
Diseases
N.A.
Drug
N.A.
Main interacting RNAs
N.A.
Moonlighting functions
TF
Localizations
N.A.
BulkPerturb-seq
Ensembl ID ENSG00000198783 Gene ID
91603 Accession
28291
Symbol
ZNF830
Alias
OMCG1;CCDC16
Full Name
zinc finger protein 830
Status
Confidence
Length
2238 bases
Strand
Plus strand
Position
17 : 34961540 - 34963777
RNA binding domain
zf-met
Summary
Predicted to enable nucleic acid binding activity and zinc ion binding activity. Predicted to be involved in several processes, including DNA-dependent DNA replication; mitotic DNA integrity checkpoint signaling; and preantral ovarian follicle growth. Predicted to act upstream of or within several processes, including blastocyst growth; chromosome organization; and intestinal epithelial structure maintenance. Predicted to be located in nucleoplasm. Predicted to be part of spliceosomal complex. Predicted to be active in nucleus. [provided by Alliance of Genome Resources, Apr 2022]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
ENSP00000354518
zf-met
PF12874 0.0011
1
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
Name
Transcript ID
bp
Protein
Translation ID
ZNF830-201
ENST00000361952
2238
372aa
ENSP00000354518
ZNF830-202
ENST00000578339
667
106aa
ENSP00000463839
Pathway ID
Pathway Name
Source
ensgID
Trait
pValue
Pubmed ID
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
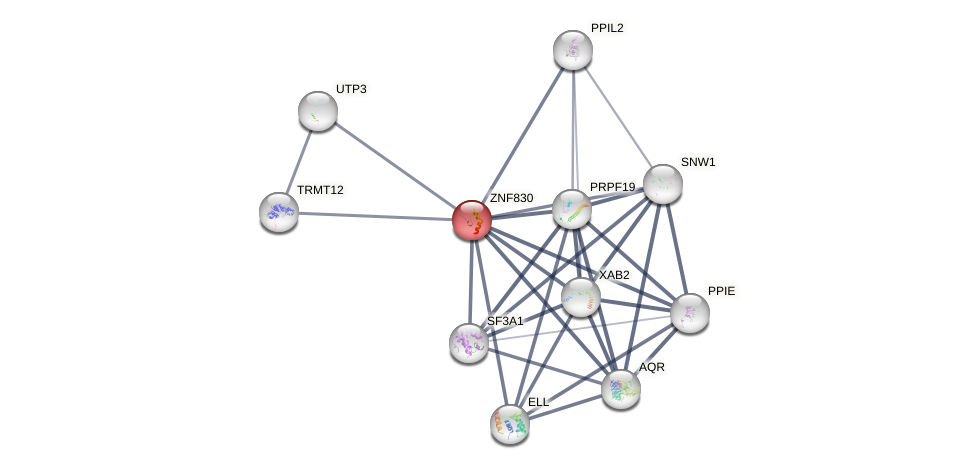
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0001541 involved_in ovarian follicle development 1 ISS Homo_sapiens(9606) Process GO:0001546 involved_in preantral ovarian follicle growth 1 ISS Homo_sapiens(9606) Process GO:0001832 involved_in blastocyst growth 1 IEA Homo_sapiens(9606) Process GO:0003676 enables nucleic acid binding 1 IEA Homo_sapiens(9606) Function GO:0005515 enables protein binding 1 IPI Homo_sapiens(9606) Function GO:0005634 is_active_in nucleus 1 IBA Homo_sapiens(9606) Component GO:0005654 located_in nucleoplasm 1 TAS Homo_sapiens(9606) Component GO:0005681 part_of spliceosomal complex 1 IEA Homo_sapiens(9606) Component GO:0005694 located_in chromosome 1 IEA Homo_sapiens(9606) Component GO:0006396 involved_in RNA processing 1 ISS Homo_sapiens(9606) Process GO:0006397 involved_in mRNA processing 1 IEA Homo_sapiens(9606) Process GO:0008270 enables zinc ion binding 1 IEA Homo_sapiens(9606) Function GO:0008380 involved_in RNA splicing 1 IEA Homo_sapiens(9606) Process GO:0016607 located_in nuclear speck 1 IEA Homo_sapiens(9606) Component GO:0033260 involved_in nuclear DNA replication 1 IBA Homo_sapiens(9606) Process GO:0033314 involved_in mitotic DNA replication checkpoint signaling 2 IBA,ISS Homo_sapiens(9606) Process GO:0043066 involved_in negative regulation of apoptotic process 1 ISS Homo_sapiens(9606) Process GO:0044773 involved_in mitotic DNA damage checkpoint signaling 1 IBA Homo_sapiens(9606) Process GO:0051276 involved_in chromosome organization 1 IEA Homo_sapiens(9606) Process GO:0051301 involved_in cell division 1 IEA Homo_sapiens(9606) Process GO:0060729 involved_in intestinal epithelial structure maintenance 1 IEA Homo_sapiens(9606) Process
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.