RBP Type
Canonical_RBPs
Diseases
SARS-COV-OC43 Flaviviruses SARS-COV-2 Dengue Zika
Drug
N.A.
Main interacting RNAs
mRNA
Moonlighting functions
N.A.
Localizations
Stress granucle
BulkPerturb-seq
N.A.
Ensembl ID ENSG00000198563 Gene ID
7919 Accession
13917
Symbol
DDX39B
Alias
BAT1;UAP56;D6S81E
Full Name
DExD-box helicase 39B
Status
Confidence
Length
12230 bases
Strand
Minus strand
Position
6 : 31530219 - 31542448
RNA binding domain
ResIII , DEAD
Summary
This gene encodes a member of the DEAD box family of RNA-dependent ATPases that mediate ATP hydrolysis during pre-mRNA splicing. The encoded protein is an essential splicing factor required for association of U2 small nuclear ribonucleoprotein with pre-mRNA, and it also plays an important role in mRNA export from the nucleus to the cytoplasm. This gene belongs to a cluster of genes localized in the vicinity of the genes encoding tumor necrosis factor alpha and tumor necrosis factor beta. These genes are all within the human major histocompatibility complex class III region. Mutations in this gene may be associated with rheumatoid arthritis. Alternative splicing results in multiple transcript variants. Related pseudogenes have been identified on both chromosomes 6 and 11. Read-through transcription also occurs between this gene and the upstream ATP6V1G2 (ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2) gene. [provided by RefSeq, Feb 2011]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
ENSP00000392672
DEAD
PF00270 8.5e-39
1
ENSP00000392672
ResIII
PF04851 1.7e-07
2
ENSP00000399371
DEAD
PF00270 1.4e-38
1
ENSP00000399371
ResIII
PF04851 2.5e-07
2
ENSP00000410313
DEAD
PF00270 1.4e-38
1
ENSP00000410313
ResIII
PF04851 1.6e-07
2
ENSP00000379475
DEAD
PF00270 2.8e-38
1
ENSP00000379475
ResIII
PF04851 5.4e-07
2
ENSP00000416269
DEAD
PF00270 2.8e-38
1
ENSP00000416269
ResIII
PF04851 5.4e-07
2
ENSP00000365347
DEAD
PF00270 3.1e-38
1
ENSP00000365347
ResIII
PF04851 5.6e-07
2
ENSP00000416350
DEAD
PF00270 1.1e-37
1
ENSP00000416350
ResIII
PF04851 3.2e-07
2
ENSP00000408000
DEAD
PF00270 7.7e-19
1
ENSP00000409426
DEAD
PF00270 5.7e-16
1
ENSP00000405707
DEAD
PF00270 2.3e-15
1
ENSP00000393984
DEAD
PF00270 3.5e-09
1
ENSP00000399841
DEAD
PF00270 1.8e-07
1
ENSP00000405245
DEAD
PF00270 3.2e-07
1
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
37261960 The RNA helicase DDX39B activates FOXP3 RNA splicing to control T regulatory cell fate.
Minato Hirano
2023-06-01
eLife
32209106 DDX39B interacts with the pattern recognition receptor pathway to inhibit NF-κB and sensitize to alkylating chemotherapy.
J Szymon Szymura
2020-03-24
BMC biology
36084138 TREX (transcription/export)-NP complex exerts a dual effect on regulating polymerase activity and replication of influenza A virus.
Lingcai Zhao
2022-09-01
PLoS pathogens
37578863 Structural basis for high-order complex of SARNP and DDX39B to facilitate mRNP assembly.
Yihu Xie
2023-08-29
Cell reports
33436563 The DDX39B/FUT3/TGFβR-I axis promotes tumor metastasis and EMT in colorectal cancer.
Chengcheng He
2021-01-12
Cell death & disease
29849633 SNP Analysis of Caries and Initial Caries in Finnish Adolescents.
Teija Raivisto
2018-01-01
International journal of dentistry
33941617 The Mammalian Ecdysoneless Protein Interacts with RNA Helicase DDX39A To Regulate Nuclear mRNA Export.
Irfana Saleem
2021-06-23
Molecular and cellular biology
35866610 A role for the Saccharomyces cerevisiae Rtt109 histone acetyltransferase in R-loop homeostasis and associated genome instability.
Carlos Juan Cañas
2022-08-30
Genetics
Name
Transcript ID
bp
Protein
Translation ID
Pathway ID
Pathway Name
Source
ensgID
Trait
pValue
Pubmed ID
12541 Breast Neoplasms 2.459E-13 17660530 12994 Diabetes Mellitus, Type 1 5.29E-68 19430480 16222 Behcet Syndrome 4.55942540272892E-05 20622878 16228 Behcet Syndrome 5.97381407474203E-06 20622878 16242 Behcet Syndrome 8.82169288626833E-07 20622878 16282 Behcet Syndrome 8.71917049005456E-07 20622878 16284 Behcet Syndrome 1.03078253218308E-06 20622878 16286 Behcet Syndrome 8.82169288626833E-07 20622878 16292 Behcet Syndrome 1.0729589781576E-06 20622878 16295 Behcet Syndrome 6.22624609680527E-07 20622878 37650 Platelet Function Tests 8.1800000E-006 - 67762 Stevens-Johnson Syndrome 2E-8 21912425 89903 Dermatitis, Atopic 2E-6 22197932
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
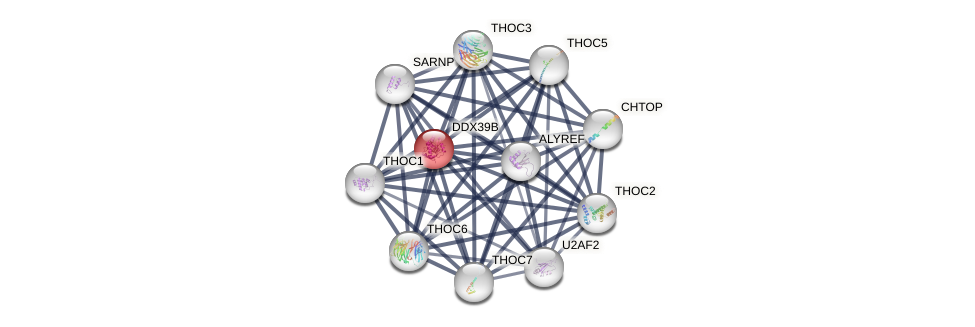
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
ENSG00000198563 homo_sapiens ENSP00000385536 28.3951 16.0494 homo_sapiens ENSP00000379475 48.3645 27.3364 ENSG00000198563 homo_sapiens ENSP00000263239 30.7463 16.5672 homo_sapiens ENSP00000379475 48.1308 25.9346 ENSG00000198563 homo_sapiens ENSP00000497641 52.0681 32.8467 homo_sapiens ENSP00000379475 50 31.5421 ENSG00000198563 homo_sapiens ENSP00000482680 26.9281 15.5556 homo_sapiens ENSP00000379475 48.1308 27.8037 ENSG00000198563 homo_sapiens ENSP00000293831 53.9409 35.2217 homo_sapiens ENSP00000379475 51.1682 33.4112 ENSG00000198563 homo_sapiens ENSP00000306117 43.0962 26.569 homo_sapiens ENSP00000379475 48.1308 29.6729 ENSG00000198563 homo_sapiens ENSP00000288071 43.0063 26.7223 homo_sapiens ENSP00000379475 48.1308 29.9065 ENSG00000198563 homo_sapiens ENSP00000481495 23.9814 12.4563 homo_sapiens ENSP00000379475 48.1308 25 ENSG00000198563 homo_sapiens ENSP00000416534 19.9612 11.2403 homo_sapiens ENSP00000379475 48.1308 27.1028 ENSG00000198563 homo_sapiens ENSP00000361232 25.067 13.5389 homo_sapiens ENSP00000379475 43.6916 23.5981 ENSG00000198563 homo_sapiens ENSP00000492643 13.9098 7.74436 homo_sapiens ENSP00000379475 43.2243 24.0654 ENSG00000198563 homo_sapiens ENSP00000238146 30.8333 16.5 homo_sapiens ENSP00000379475 43.2243 23.1308 ENSG00000198563 homo_sapiens ENSP00000225792 34.6906 19.2182 homo_sapiens ENSP00000379475 49.7664 27.5701 ENSG00000198563 homo_sapiens ENSP00000368667 32.9636 17.1157 homo_sapiens ENSP00000379475 48.5981 25.2336 ENSG00000198563 homo_sapiens ENSP00000330460 32.4717 17.609 homo_sapiens ENSP00000379475 46.9626 25.4673 ENSG00000198563 homo_sapiens ENSP00000263576 40.9938 23.3954 homo_sapiens ENSP00000379475 46.2617 26.4019 ENSG00000198563 homo_sapiens ENSP00000380495 28.8288 16.8168 homo_sapiens ENSP00000379475 44.8598 26.1682 ENSG00000198563 homo_sapiens ENSP00000314348 22.4 12 homo_sapiens ENSP00000379475 45.7944 24.5327 ENSG00000198563 homo_sapiens ENSP00000346120 27.9693 15.198 homo_sapiens ENSP00000379475 51.1682 27.8037 ENSG00000198563 homo_sapiens ENSP00000374574 21.0021 12.58 homo_sapiens ENSP00000379475 46.028 27.5701 ENSG00000198563 homo_sapiens ENSP00000358716 23.0583 15.534 homo_sapiens ENSP00000379475 44.3925 29.9065 ENSG00000198563 homo_sapiens ENSP00000442266 40.5797 24.8447 homo_sapiens ENSP00000379475 45.7944 28.0374 ENSG00000198563 homo_sapiens ENSP00000362687 29.1723 14.9254 homo_sapiens ENSP00000379475 50.2336 25.7009 ENSG00000198563 homo_sapiens ENSP00000479504 33.0551 19.5326 homo_sapiens ENSP00000379475 46.2617 27.3364 ENSG00000198563 homo_sapiens ENSP00000359361 33.4877 17.7469 homo_sapiens ENSP00000379475 50.7009 26.8692 ENSG00000198563 homo_sapiens ENSP00000326381 54.0541 35.6265 homo_sapiens ENSP00000379475 51.4019 33.8785 ENSG00000198563 homo_sapiens ENSP00000258772 34.3693 19.0128 homo_sapiens ENSP00000379475 43.9252 24.2991 ENSG00000198563 homo_sapiens ENSP00000494040 31.8731 18.7311 homo_sapiens ENSP00000379475 49.2991 28.972 ENSG00000198563 homo_sapiens ENSP00000336725 31.5152 18.6364 homo_sapiens ENSP00000379475 48.5981 28.7383 ENSG00000198563 homo_sapiens ENSP00000247003 40.5797 24.8447 homo_sapiens ENSP00000379475 45.7944 28.0374 ENSG00000198563 homo_sapiens ENSP00000330349 33.119 18.9711 homo_sapiens ENSP00000379475 48.1308 27.5701 ENSG00000198563 homo_sapiens ENSP00000242776 95.7845 89.2272 homo_sapiens ENSP00000379475 95.5607 89.0187 ENSG00000198563 homo_sapiens ENSP00000233084 29.0541 15.2703 homo_sapiens ENSP00000379475 50.2336 26.4019 ENSG00000198563 homo_sapiens ENSP00000304072 24.2906 13.6209 homo_sapiens ENSP00000379475 50 28.0374 ENSG00000198563 homo_sapiens ENSP00000350698 42.8571 26.3736 homo_sapiens ENSP00000379475 45.5607 28.0374 ENSG00000198563 homo_sapiens ENSP00000310723 25.3659 14.6341 homo_sapiens ENSP00000379475 48.5981 28.0374 ENSG00000198563 homo_sapiens ENSP00000332340 32.4074 19.6296 homo_sapiens ENSP00000379475 40.8879 24.7664 ENSG00000198563 homo_sapiens ENSP00000424838 28.3149 16.4365 homo_sapiens ENSP00000379475 47.8972 27.8037 ENSG00000198563 homo_sapiens ENSP00000390999 100 100 homo_sapiens ENSP00000379475 100 100 ENSG00000198563 homo_sapiens ENSP00000405560 100 100 homo_sapiens ENSP00000379475 100 100 ENSG00000198563 homo_sapiens ENSP00000387994 100 100 homo_sapiens ENSP00000379475 100 100 ENSG00000198563 homo_sapiens ENSP00000399542 99.6951 99.6951 homo_sapiens ENSP00000379475 76.4019 76.4019 ENSG00000198563 homo_sapiens ENSP00000391463 100 100 homo_sapiens ENSP00000379475 100 100 ENSG00000198563 homo_sapiens ENSP00000399030 100 100 homo_sapiens ENSP00000379475 100 100 ENSG00000198563 homo_sapiens ENSP00000383152 100 100 homo_sapiens ENSP00000379475 100 100
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
ENSG00000198563 homo_sapiens ENSP00000379475 82.4766 82.4766 nomascus_leucogenys ENSNLEP00000007389 99.7175 99.7175 ENSG00000198563 homo_sapiens ENSP00000379475 100 100 pan_paniscus ENSPPAP00000040882 100 100 ENSG00000198563 homo_sapiens ENSP00000379475 100 100 pongo_abelii ENSPPYP00000018374 100 100 ENSG00000198563 homo_sapiens ENSP00000379475 100 100 pan_troglodytes ENSPTRP00000054521 100 100 ENSG00000198563 homo_sapiens ENSP00000379475 100 100 gorilla_gorilla ENSGGOP00000005862 100 100 ENSG00000198563 homo_sapiens ENSP00000379475 99.0654 99.0654 ornithorhynchus_anatinus ENSOANP00000008158 94.0133 94.0133 ENSG00000198563 homo_sapiens ENSP00000379475 99.2991 99.0654 sarcophilus_harrisii ENSSHAP00000033244 98.1524 97.9215 ENSG00000198563 homo_sapiens ENSP00000379475 85.2804 85.0467 notamacropus_eugenii ENSMEUP00000007544 85.2804 85.0467 ENSG00000198563 homo_sapiens ENSP00000379475 100 100 dasypus_novemcinctus ENSDNOP00000007910 100 100 ENSG00000198563 homo_sapiens ENSP00000379475 53.0374 52.8037 erinaceus_europaeus ENSEEUP00000010964 72.524 72.2045 ENSG00000198563 homo_sapiens ENSP00000379475 78.0374 76.8692 echinops_telfairi ENSETEP00000007840 78.0374 76.8692 ENSG00000198563 homo_sapiens ENSP00000379475 99.2991 98.8318 callithrix_jacchus ENSCJAP00000079502 97.4771 97.0183 ENSG00000198563 homo_sapiens ENSP00000379475 83.6449 81.5421 cercocebus_atys ENSCATP00000042163 84.2353 82.1176 ENSG00000198563 homo_sapiens ENSP00000379475 100 100 macaca_fascicularis ENSMFAP00000035785 100 100 ENSG00000198563 homo_sapiens ENSP00000379475 100 100 macaca_mulatta ENSMMUP00000050210 100 100 ENSG00000198563 homo_sapiens ENSP00000379475 100 100 macaca_nemestrina ENSMNEP00000035839 100 100 ENSG00000198563 homo_sapiens ENSP00000379475 100 100 mandrillus_leucophaeus ENSMLEP00000039530 100 100 ENSG00000198563 homo_sapiens ENSP00000379475 92.5234 92.5234 tursiops_truncatus ENSTTRP00000013346 91.6667 91.6667 ENSG00000198563 homo_sapiens ENSP00000379475 100 99.5327 vulpes_vulpes ENSVVUP00000027896 92.4406 92.0086 ENSG00000198563 homo_sapiens ENSP00000379475 99.5327 99.5327 mus_spretus MGP_SPRETEiJ_P0046399 99.5327 99.5327 ENSG00000198563 homo_sapiens ENSP00000379475 100 100 equus_asinus ENSEASP00005022353 100 100 ENSG00000198563 homo_sapiens ENSP00000379475 100 99.7664 capra_hircus ENSCHIP00000002143 100 99.7664 ENSG00000198563 homo_sapiens ENSP00000379475 99.7664 99.7664 loxodonta_africana ENSLAFP00000011893 100 100 ENSG00000198563 homo_sapiens ENSP00000379475 100 99.7664 panthera_leo ENSPLOP00000012091 100 99.7664 ENSG00000198563 homo_sapiens ENSP00000379475 100 99.7664 ailuropoda_melanoleuca ENSAMEP00000009997 100 99.7664 ENSG00000198563 homo_sapiens ENSP00000379475 100 99.7664 bos_indicus_hybrid ENSBIXP00005035691 100 99.7664 ENSG00000198563 homo_sapiens ENSP00000379475 99.7664 99.5327 oryctolagus_cuniculus ENSOCUP00000025395 100 99.7658 ENSG00000198563 homo_sapiens ENSP00000379475 99.2991 99.0654 equus_caballus ENSECAP00000052685 94.8661 94.6429 ENSG00000198563 homo_sapiens ENSP00000379475 100 99.5327 canis_lupus_familiaris ENSCAFP00845014141 100 99.5327 ENSG00000198563 homo_sapiens ENSP00000379475 100 99.7664 panthera_pardus ENSPPRP00000006435 100 99.7664 ENSG00000198563 homo_sapiens ENSP00000379475 99.7664 99.5327 marmota_marmota_marmota ENSMMMP00000010373 99.7664 99.5327 ENSG00000198563 homo_sapiens ENSP00000379475 100 100 balaenoptera_musculus ENSBMSP00010021167 100 100 ENSG00000198563 homo_sapiens ENSP00000379475 99.7664 99.7664 cavia_porcellus ENSCPOP00000002121 99.7664 99.7664 ENSG00000198563 homo_sapiens ENSP00000379475 100 99.7664 felis_catus ENSFCAP00000028032 100 99.7664 ENSG00000198563 homo_sapiens ENSP00000379475 88.5514 86.9159 ursus_americanus ENSUAMP00000036056 91.9903 90.2913 ENSG00000198563 homo_sapiens ENSP00000379475 99.2991 98.8318 monodelphis_domestica ENSMODP00000019700 92.3913 91.9565 ENSG00000198563 homo_sapiens ENSP00000379475 99.2991 99.0654 bison_bison_bison ENSBBBP00000023594 91.0064 90.7923 ENSG00000198563 homo_sapiens ENSP00000379475 100 100 monodon_monoceros ENSMMNP00015016942 100 100 ENSG00000198563 homo_sapiens ENSP00000379475 97.1963 96.4953 sus_scrofa ENSSSCP00000031099 88.5106 87.8723 ENSG00000198563 homo_sapiens ENSP00000379475 100 100 microcebus_murinus ENSMICP00000015802 100 100 ENSG00000198563 homo_sapiens ENSP00000379475 99.5327 99.5327 octodon_degus ENSODEP00000024705 99.5327 99.5327 ENSG00000198563 homo_sapiens ENSP00000379475 99.7664 99.2991 jaculus_jaculus ENSJJAP00000003018 99.7664 99.2991 ENSG00000198563 homo_sapiens ENSP00000379475 100 99.7664 ovis_aries_rambouillet ENSOARP00020020927 100 99.7664 ENSG00000198563 homo_sapiens ENSP00000379475 91.8224 90.8878 ursus_maritimus ENSUMAP00000020768 97.037 96.0494 ENSG00000198563 homo_sapiens ENSP00000379475 100 99.7664 ochotona_princeps ENSOPRP00000010876 100 99.7664 ENSG00000198563 homo_sapiens ENSP00000379475 100 100 delphinapterus_leucas ENSDLEP00000026948 88.9813 88.9813 ENSG00000198563 homo_sapiens ENSP00000379475 100 100 physeter_catodon ENSPCTP00005019198 100 100 ENSG00000198563 homo_sapiens ENSP00000379475 42.0561 40.8879 mus_caroli MGP_CAROLIEiJ_P0044498 97.8261 95.1087 ENSG00000198563 homo_sapiens ENSP00000379475 99.5327 99.5327 mus_musculus ENSMUSP00000133428 99.5327 99.5327 ENSG00000198563 homo_sapiens ENSP00000379475 87.6168 84.8131 dipodomys_ordii ENSDORP00000004664 93.985 90.9774 ENSG00000198563 homo_sapiens ENSP00000379475 100 99.7664 mustela_putorius_furo ENSMPUP00000011315 100 99.7664 ENSG00000198563 homo_sapiens ENSP00000379475 100 100 aotus_nancymaae ENSANAP00000025459 100 100 ENSG00000198563 homo_sapiens ENSP00000379475 34.3458 33.6449 saimiri_boliviensis_boliviensis ENSSBOP00000028168 97.351 95.3642 ENSG00000198563 homo_sapiens ENSP00000379475 89.7196 89.7196 saimiri_boliviensis_boliviensis ENSSBOP00000038921 100 100 ENSG00000198563 homo_sapiens ENSP00000379475 50.7009 50.7009 vicugna_pacos ENSVPAP00000008533 60.9551 60.9551 ENSG00000198563 homo_sapiens ENSP00000379475 99.0654 99.0654 chlorocebus_sabaeus ENSCSAP00000004768 99.7647 99.7647 ENSG00000198563 homo_sapiens ENSP00000379475 77.5701 76.8692 tupaia_belangeri ENSTBEP00000003234 78.3019 77.5943 ENSG00000198563 homo_sapiens ENSP00000379475 99.7664 99.2991 mesocricetus_auratus ENSMAUP00000005984 99.7664 99.2991 ENSG00000198563 homo_sapiens ENSP00000379475 100 100 phocoena_sinus ENSPSNP00000013714 100 100 ENSG00000198563 homo_sapiens ENSP00000379475 100 100 camelus_dromedarius ENSCDRP00005029377 100 100 ENSG00000198563 homo_sapiens ENSP00000379475 100 100 carlito_syrichta ENSTSYP00000028763 100 100 ENSG00000198563 homo_sapiens ENSP00000379475 99.0654 98.8318 panthera_tigris_altaica ENSPTIP00000020071 99.0654 98.8318 ENSG00000198563 homo_sapiens ENSP00000379475 99.0654 99.0654 rattus_norvegicus ENSRNOP00000093667 92.9825 92.9825 ENSG00000198563 homo_sapiens ENSP00000379475 100 99.7664 prolemur_simus ENSPSMP00000025723 99.7669 99.5338 ENSG00000198563 homo_sapiens ENSP00000379475 99.7664 99.5327 microtus_ochrogaster ENSMOCP00000010919 99.7664 99.5327 ENSG00000198563 homo_sapiens ENSP00000379475 76.8692 76.6355 sorex_araneus ENSSARP00000004741 87.9679 87.7005 ENSG00000198563 homo_sapiens ENSP00000379475 99.5327 99.2991 peromyscus_maniculatus_bairdii ENSPEMP00000018246 99.5327 99.2991 ENSG00000198563 homo_sapiens ENSP00000379475 100 99.7664 bos_mutus ENSBMUP00000017910 100 99.7664 ENSG00000198563 homo_sapiens ENSP00000379475 63.785 61.215 mus_spicilegus ENSMSIP00000004714 84.2593 80.8642 ENSG00000198563 homo_sapiens ENSP00000379475 60.0467 58.8785 cricetulus_griseus_chok1gshd ENSCGRP00001017226 92.1147 90.3226 ENSG00000198563 homo_sapiens ENSP00000379475 99.5327 99.5327 pteropus_vampyrus ENSPVAP00000004405 99.5327 99.5327 ENSG00000198563 homo_sapiens ENSP00000379475 72.6636 72.6636 myotis_lucifugus ENSMLUP00000001542 100 100 ENSG00000198563 homo_sapiens ENSP00000379475 100 100 vombatus_ursinus ENSVURP00010002353 100 100 ENSG00000198563 homo_sapiens ENSP00000379475 98.8318 98.3645 rhinolophus_ferrumequinum ENSRFEP00010001850 79.2135 78.839 ENSG00000198563 homo_sapiens ENSP00000379475 100 99.7664 neovison_vison ENSNVIP00000021209 100 99.7664 ENSG00000198563 homo_sapiens ENSP00000379475 100 99.7664 bos_taurus ENSBTAP00000067484 100 99.7664 ENSG00000198563 homo_sapiens ENSP00000379475 87.3832 85.0467 heterocephalus_glaber_female ENSHGLP00000039438 80.2575 78.1116 ENSG00000198563 homo_sapiens ENSP00000379475 89.7196 89.7196 rhinopithecus_bieti ENSRBIP00000014406 100 100 ENSG00000198563 homo_sapiens ENSP00000379475 100 99.5327 canis_lupus_dingo ENSCAFP00020026149 100 99.5327 ENSG00000198563 homo_sapiens ENSP00000379475 90.8878 89.7196 sciurus_vulgaris ENSSVLP00005005587 97.7387 96.4824 ENSG00000198563 homo_sapiens ENSP00000379475 100 100 phascolarctos_cinereus ENSPCIP00000040712 100 100 ENSG00000198563 homo_sapiens ENSP00000379475 66.1215 66.1215 procavia_capensis ENSPCAP00000003265 100 100 ENSG00000198563 homo_sapiens ENSP00000379475 99.0654 98.8318 nannospalax_galili ENSNGAP00000024809 94.6429 94.4196 ENSG00000198563 homo_sapiens ENSP00000379475 100 99.7664 bos_grunniens ENSBGRP00000020200 100 99.7664 ENSG00000198563 homo_sapiens ENSP00000379475 99.7664 99.7664 chinchilla_lanigera ENSCLAP00000021256 99.7664 99.7664 ENSG00000198563 homo_sapiens ENSP00000379475 91.3551 89.2523 cervus_hanglu_yarkandensis ENSCHYP00000007685 88.4615 86.4253 ENSG00000198563 homo_sapiens ENSP00000379475 100 100 catagonus_wagneri ENSCWAP00000007204 100 100 ENSG00000198563 homo_sapiens ENSP00000379475 99.7664 99.5327 ictidomys_tridecemlineatus ENSSTOP00000011504 99.7664 99.5327 ENSG00000198563 homo_sapiens ENSP00000379475 99.5327 98.8318 otolemur_garnettii ENSOGAP00000019155 99.3007 98.6014 ENSG00000198563 homo_sapiens ENSP00000379475 100 100 cebus_imitator ENSCCAP00000020179 100 100 ENSG00000198563 homo_sapiens ENSP00000379475 98.8318 98.5981 urocitellus_parryii ENSUPAP00010026091 97.0183 96.789 ENSG00000198563 homo_sapiens ENSP00000379475 100 99.7664 moschus_moschiferus ENSMMSP00000023756 100 99.7664 ENSG00000198563 homo_sapiens ENSP00000379475 100 100 rhinopithecus_roxellana ENSRROP00000031674 100 100 ENSG00000198563 homo_sapiens ENSP00000379475 99.5327 99.5327 mus_pahari MGP_PahariEiJ_P0044037 99.5327 99.5327 ENSG00000198563 homo_sapiens ENSP00000379475 100 100 propithecus_coquereli ENSPCOP00000013989 100 100 ENSG00000198563 homo_sapiens ENSP00000379475 92.9907 91.1215 latimeria_chalumnae ENSLACP00000002361 91.9169 90.0693 ENSG00000198563 homo_sapiens ENSP00000379475 86.9159 84.1122 clupea_harengus ENSCHAP00000036871 83.0357 80.3571 ENSG00000198563 homo_sapiens ENSP00000379475 96.729 93.9252 danio_rerio ENSDARP00000052354 95.1724 92.4138 ENSG00000198563 homo_sapiens ENSP00000379475 96.4953 93.4579 carassius_auratus ENSCARP00000100704 87.5 84.7458 ENSG00000198563 homo_sapiens ENSP00000379475 96.028 93.4579 astyanax_mexicanus ENSAMXP00000018374 94.2661 91.7431 ENSG00000198563 homo_sapiens ENSP00000379475 92.0561 88.0841 ictalurus_punctatus ENSIPUP00000023394 84.188 80.5556 ENSG00000198563 homo_sapiens ENSP00000379475 95.3271 92.2897 esox_lucius ENSELUP00000043949 90.0662 87.1965 ENSG00000198563 homo_sapiens ENSP00000379475 96.028 92.9907 electrophorus_electricus ENSEEEP00000039337 88.7689 85.9611 ENSG00000198563 homo_sapiens ENSP00000379475 94.8598 92.0561 oncorhynchus_kisutch ENSOKIP00005076058 90.2222 87.5556 ENSG00000198563 homo_sapiens ENSP00000379475 94.8598 92.0561 oncorhynchus_mykiss ENSOMYP00000138995 81.5261 79.1165 ENSG00000198563 homo_sapiens ENSP00000379475 95.3271 92.5234 salmo_salar ENSSSAP00000158728 87.1795 84.6154 ENSG00000198563 homo_sapiens ENSP00000379475 94.6262 91.5888 salmo_trutta ENSSTUP00000036474 90.4018 87.5 ENSG00000198563 homo_sapiens ENSP00000379475 95.0935 92.5234 oreochromis_niloticus ENSONIP00000028282 89.4505 87.033 ENSG00000198563 homo_sapiens ENSP00000379475 49.2991 46.9626 haplochromis_burtoni ENSHBUP00000022244 93.7778 89.3333 ENSG00000198563 homo_sapiens ENSP00000379475 94.3925 92.0561 astatotilapia_calliptera ENSACLP00000027646 92.4485 90.1602 ENSG00000198563 homo_sapiens ENSP00000379475 96.028 93.2243 lates_calcarifer ENSLCAP00010020371 89.5425 86.9281 ENSG00000198563 homo_sapiens ENSP00000379475 96.9626 94.1589 xenopus_tropicalis ENSXETP00000086329 96.7366 93.9394 ENSG00000198563 homo_sapiens ENSP00000379475 63.785 63.785 chrysemys_picta_bellii ENSCPBP00000020397 100 100 ENSG00000198563 homo_sapiens ENSP00000379475 98.5981 97.8972 sphenodon_punctatus ENSSPUP00000016565 98.5981 97.8972 ENSG00000198563 homo_sapiens ENSP00000379475 97.1963 95.7944 laticauda_laticaudata ENSLLTP00000022232 97.8824 96.4706 ENSG00000198563 homo_sapiens ENSP00000379475 98.1308 96.729 notechis_scutatus ENSNSUP00000012850 98.1308 96.729 ENSG00000198563 homo_sapiens ENSP00000379475 98.1308 96.729 pseudonaja_textilis ENSPTXP00000025264 98.1308 96.729 ENSG00000198563 homo_sapiens ENSP00000379475 98.5981 97.4299 anolis_carolinensis ENSACAP00000012097 93.7778 92.6667 ENSG00000198563 homo_sapiens ENSP00000379475 47.8972 41.3551 neolamprologus_brichardi ENSNBRP00000001327 62.1212 53.6364 ENSG00000198563 homo_sapiens ENSP00000379475 97.8972 96.4953 naja_naja ENSNNAP00000003425 98.1265 96.7213 ENSG00000198563 homo_sapiens ENSP00000379475 98.3645 97.6636 podarcis_muralis ENSPMRP00000004389 94.1834 93.5123 ENSG00000198563 homo_sapiens ENSP00000379475 98.3645 97.1963 erpetoichthys_calabaricus ENSECRP00000000630 98.3645 97.1963 ENSG00000198563 homo_sapiens ENSP00000379475 98.1308 96.729 salvator_merianae ENSSMRP00000020802 98.1308 96.729 ENSG00000198563 homo_sapiens ENSP00000379475 95.3271 93.4579 amphilophus_citrinellus ENSACIP00000005755 94.0092 92.1659 ENSG00000198563 homo_sapiens ENSP00000379475 98.5981 98.1308 gopherus_evgoodei ENSGEVP00005019930 98.5981 98.1308 ENSG00000198563 homo_sapiens ENSP00000379475 80.1402 76.4019 chelonoidis_abingdonii ENSCABP00000031545 62.3636 59.4545 ENSG00000198563 homo_sapiens ENSP00000379475 96.2617 94.3925 seriola_dumerili ENSSDUP00000015851 95.3704 93.5185 ENSG00000198563 homo_sapiens ENSP00000379475 96.028 93.9252 myripristis_murdjan ENSMMDP00005011534 95.1389 93.0556 ENSG00000198563 homo_sapiens ENSP00000379475 46.729 46.2617 terrapene_carolina_triunguis ENSTMTP00000018555 92.5926 91.6667 ENSG00000198563 homo_sapiens ENSP00000379475 46.729 46.2617 terrapene_carolina_triunguis ENSTMTP00000018606 92.5926 91.6667 ENSG00000198563 homo_sapiens ENSP00000379475 95.5607 92.2897 hucho_hucho ENSHHUP00000037251 91.704 88.565 ENSG00000198563 homo_sapiens ENSP00000379475 96.4953 93.9252 pygocentrus_nattereri ENSPNAP00000036253 94.7248 92.2018 ENSG00000198563 homo_sapiens ENSP00000379475 95.7944 92.9907 anabas_testudineus ENSATEP00000065477 90.708 88.0531 ENSG00000198563 homo_sapiens ENSP00000379475 96.4953 94.1589 seriola_lalandi_dorsalis ENSSLDP00000020154 95.6019 93.287 ENSG00000198563 homo_sapiens ENSP00000379475 85.0467 78.0374 labrus_bergylta ENSLBEP00000015818 83.6782 76.7816 ENSG00000198563 homo_sapiens ENSP00000379475 63.0841 55.1402 labrus_bergylta ENSLBEP00000022371 77.3639 67.6218 ENSG00000198563 homo_sapiens ENSP00000379475 85.0467 82.4766 pundamilia_nyererei ENSPNYP00000013717 89.2157 86.5196 ENSG00000198563 homo_sapiens ENSP00000379475 89.2523 84.8131 denticeps_clupeoides ENSDCDP00000012670 83.7719 79.6053 ENSG00000198563 homo_sapiens ENSP00000379475 96.028 92.757 denticeps_clupeoides ENSDCDP00000044919 79.4971 76.7892 ENSG00000198563 homo_sapiens ENSP00000379475 95.7944 93.6916 larimichthys_crocea ENSLCRP00005007906 88.3621 86.4224 ENSG00000198563 homo_sapiens ENSP00000379475 22.1963 17.5234 ficedula_albicollis ENSFALP00000030085 53.6723 42.3729 ENSG00000198563 homo_sapiens ENSP00000379475 95.5607 92.5234 cyprinus_carpio_carpio ENSCCRP00000079143 93.1663 90.205 ENSG00000198563 homo_sapiens ENSP00000379475 95.7944 92.2897 sinocyclocheilus_grahami ENSSGRP00000042790 91.1111 87.7778 ENSG00000198563 homo_sapiens ENSP00000379475 96.4953 93.4579 sinocyclocheilus_grahami ENSSGRP00000025232 88.2479 85.4701 ENSG00000198563 homo_sapiens ENSP00000379475 94.6262 92.0561 maylandia_zebra ENSMZEP00005023594 92.6773 90.1602 ENSG00000198563 homo_sapiens ENSP00000379475 96.2617 92.9907 paramormyrops_kingsleyae ENSPKIP00000030493 94.2792 91.0755 ENSG00000198563 homo_sapiens ENSP00000379475 94.1589 89.7196 scleropages_formosus ENSSFOP00015078043 84.1336 80.167 ENSG00000198563 homo_sapiens ENSP00000379475 97.1963 94.8598 leptobrachium_leishanense ENSLLEP00000018130 83.3667 81.3627
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0000245 involved_in spliceosomal complex assembly 1 IDA Homo_sapiens(9606) Process GO:0000346 part_of transcription export complex 1 IDA Homo_sapiens(9606) Component GO:0000398 involved_in mRNA splicing, via spliceosome 2 IBA,IGI Homo_sapiens(9606) Process GO:0003723 enables RNA binding 2 HDA,IBA Homo_sapiens(9606) Function GO:0003724 enables RNA helicase activity 2 IBA,IDA Homo_sapiens(9606) Function GO:0005515 enables protein binding 1 IPI Homo_sapiens(9606) Function GO:0005524 enables ATP binding 1 IEA Homo_sapiens(9606) Function GO:0005634 located_in nucleus 1 IDA Homo_sapiens(9606) Component GO:0005654 located_in nucleoplasm 1 TAS Homo_sapiens(9606) Component GO:0005681 part_of spliceosomal complex 1 IDA Homo_sapiens(9606) Component GO:0005687 part_of U4 snRNP 1 IDA Homo_sapiens(9606) Component GO:0005688 part_of U6 snRNP 1 IDA Homo_sapiens(9606) Component GO:0005737 colocalizes_with cytoplasm 1 IDA Homo_sapiens(9606) Component GO:0006405 involved_in RNA export from nucleus 1 TAS Homo_sapiens(9606) Process GO:0006406 involved_in mRNA export from nucleus 3 IBA,IDA,IGI Homo_sapiens(9606) Process GO:0008186 enables ATP-dependent activity, acting on RNA 1 IDA Homo_sapiens(9606) Function GO:0008380 involved_in RNA splicing 1 IDA Homo_sapiens(9606) Process GO:0010501 involved_in RNA secondary structure unwinding 1 IDA Homo_sapiens(9606) Process GO:0016607 located_in nuclear speck 1 IDA Homo_sapiens(9606) Component GO:0016887 enables ATP hydrolysis activity 1 EXP Homo_sapiens(9606) Function GO:0017070 enables U6 snRNA binding 1 IDA Homo_sapiens(9606) Function GO:0030621 enables U4 snRNA binding 1 IDA Homo_sapiens(9606) Function GO:0032786 involved_in positive regulation of DNA-templated transcription, elongation 1 IMP Homo_sapiens(9606) Process GO:0042802 enables identical protein binding 1 IPI Homo_sapiens(9606) Function GO:0043008 enables ATP-dependent protein binding 1 IDA Homo_sapiens(9606) Function GO:0046784 involved_in viral mRNA export from host cell nucleus 1 IDA Homo_sapiens(9606) Process GO:2000002 involved_in negative regulation of DNA damage checkpoint 1 IMP Homo_sapiens(9606) Process
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.