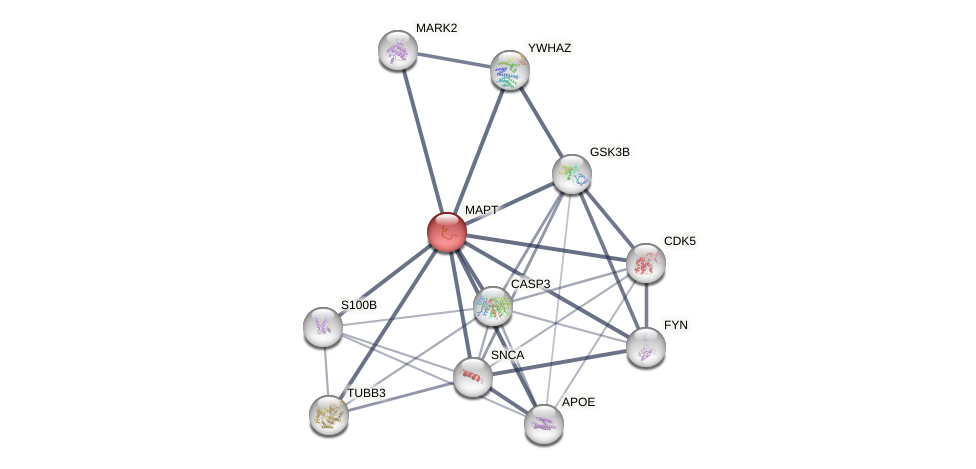
| GO:0000226 | involved_in microtubule cytoskeleton organization | 3 | IBA,IDA,NAS | Homo_sapiens(9606) | Process |
| GO:0001774 | involved_in microglial cell activation | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0003677 | enables DNA binding | 2 | ISS,TAS | Homo_sapiens(9606) | Function |
| GO:0003680 | enables minor groove of adenine-thymine-rich DNA binding | 1 | TAS | Homo_sapiens(9606) | Function |
| GO:0003690 | enables double-stranded DNA binding | 1 | TAS | Homo_sapiens(9606) | Function |
| GO:0003697 | enables single-stranded DNA binding | 1 | TAS | Homo_sapiens(9606) | Function |
| GO:0003723 | enables RNA binding | 1 | TAS | Homo_sapiens(9606) | Function |
| GO:0003779 | enables actin binding | 1 | TAS | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005576 | located_in extracellular region | 1 | NAS | Homo_sapiens(9606) | Component |
| GO:0005634 | located_in nucleus | 2 | ISS,TAS | Homo_sapiens(9606) | Component |
| GO:0005737 | located_in cytoplasm | 3 | IDA,IMP,ISS | Homo_sapiens(9606) | Component |
| GO:0005739 | located_in mitochondrion | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0005829 | located_in cytosol | 3 | IDA,ISS,TAS | Homo_sapiens(9606) | Component |
| GO:0005874 | colocalizes_with microtubule | 1 | ISS | Homo_sapiens(9606) | Component |
| GO:0005886 | located_in plasma membrane | 2 | IDA,TAS | Homo_sapiens(9606) | Component |
| GO:0006475 | involved_in internal protein amino acid acetylation | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0006919 | involved_in activation of cysteine-type endopeptidase activity involved in apoptotic process | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0006974 | involved_in DNA damage response | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0007267 | involved_in cell-cell signaling | 1 | NAS | Homo_sapiens(9606) | Process |
| GO:0007613 | involved_in memory | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0008017 | enables microtubule binding | 5 | IBA,IDA,IMP,NAS,TAS | Homo_sapiens(9606) | Function |
| GO:0010288 | involved_in response to lead ion | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0010506 | acts_upstream_of_or_within regulation of autophagy | 1 | IGI | Homo_sapiens(9606) | Process |
| GO:0010629 | involved_in negative regulation of gene expression | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0010917 | involved_in negative regulation of mitochondrial membrane potential | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0015630 | located_in microtubule cytoskeleton | 2 | IDA,NAS | Homo_sapiens(9606) | Component |
| GO:0016020 | colocalizes_with membrane | 1 | IMP | Homo_sapiens(9606) | Component |
| GO:0016072 | involved_in rRNA metabolic process | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0016607 | located_in nuclear speck | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0017124 | enables SH3 domain binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0019896 | involved_in axonal transport of mitochondrion | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0019899 | enables enzyme binding | 2 | IPI,ISS | Homo_sapiens(9606) | Function |
| GO:0019901 | enables protein kinase binding | 2 | IPI,ISS | Homo_sapiens(9606) | Function |
| GO:0021954 | involved_in central nervous system neuron development | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0030424 | located_in axon | 3 | IDA,NAS,TAS | Homo_sapiens(9606) | Component |
| GO:0030425 | located_in dendrite | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0030426 | located_in growth cone | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0030673 | located_in axolemma | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0030674 | enables protein-macromolecule adaptor activity | 1 | TAS | Homo_sapiens(9606) | Function |
| GO:0031110 | involved_in regulation of microtubule polymerization or depolymerization | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0031113 | involved_in regulation of microtubule polymerization | 2 | NAS,TAS | Homo_sapiens(9606) | Process |
| GO:0031116 | involved_in positive regulation of microtubule polymerization | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0031122 | involved_in cytoplasmic microtubule organization | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0031175 | involved_in neuron projection development | 2 | IBA,TAS | Homo_sapiens(9606) | Process |
| GO:0032930 | involved_in positive regulation of superoxide anion generation | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0033044 | involved_in regulation of chromosome organization | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0033673 | involved_in negative regulation of kinase activity | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0034063 | involved_in stress granule assembly | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0034185 | enables apolipoprotein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0034399 | located_in nuclear periphery | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0034452 | enables dynactin binding | 1 | TAS | Homo_sapiens(9606) | Function |
| GO:0034605 | involved_in cellular response to heat | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0034614 | involved_in cellular response to reactive oxygen species | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0035091 | enables phosphatidylinositol binding | 1 | TAS | Homo_sapiens(9606) | Function |
| GO:0036464 | located_in cytoplasmic ribonucleoprotein granule | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0036477 | located_in somatodendritic compartment | 2 | IMP,TAS | Homo_sapiens(9606) | Component |
| GO:0042802 | enables identical protein binding | 3 | IDA,IMP,IPI | Homo_sapiens(9606) | Function |
| GO:0043005 | is_active_in neuron projection | 1 | IBA | Homo_sapiens(9606) | Component |
| GO:0043025 | located_in neuronal cell body | 1 | IMP | Homo_sapiens(9606) | Component |
| GO:0043197 | located_in dendritic spine | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0043565 | enables sequence-specific DNA binding | 1 | TAS | Homo_sapiens(9606) | Function |
| GO:0044297 | located_in cell body | 2 | IDA,ISS | Homo_sapiens(9606) | Component |
| GO:0044304 | located_in main axon | 1 | ISS | Homo_sapiens(9606) | Component |
| GO:0045121 | located_in membrane raft | 1 | ISS | Homo_sapiens(9606) | Component |
| GO:0045298 | part_of tubulin complex | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0045773 | involved_in positive regulation of axon extension | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0046785 | involved_in microtubule polymerization | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0048143 | involved_in astrocyte activation | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0048167 | involved_in regulation of synaptic plasticity | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0048312 | involved_in intracellular distribution of mitochondria | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0048699 | involved_in generation of neurons | 1 | NAS | Homo_sapiens(9606) | Process |
| GO:0050808 | involved_in synapse organization | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0050848 | involved_in regulation of calcium-mediated signaling | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0051087 | enables protein-folding chaperone binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0051258 | involved_in protein polymerization | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0051721 | enables protein phosphatase 2A binding | 1 | TAS | Homo_sapiens(9606) | Function |
| GO:0051879 | enables Hsp90 protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0061564 | involved_in axon development | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0070507 | involved_in regulation of microtubule cytoskeleton organization | 2 | IMP,TAS | Homo_sapiens(9606) | Process |
| GO:0071813 | enables lipoprotein particle binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0072386 | involved_in plus-end-directed organelle transport along microtubule | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0090140 | involved_in regulation of mitochondrial fission | 1 | IC | Homo_sapiens(9606) | Process |
| GO:0090258 | involved_in negative regulation of mitochondrial fission | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0097386 | located_in glial cell projection | 1 | ISS | Homo_sapiens(9606) | Component |
| GO:0097418 | located_in neurofibrillary tangle | 2 | IDA,NAS | Homo_sapiens(9606) | Component |
| GO:0097435 | involved_in supramolecular fiber organization | 2 | IDA,IMP | Homo_sapiens(9606) | Process |
| GO:0098930 | involved_in axonal transport | 2 | NAS,TAS | Homo_sapiens(9606) | Process |
| GO:0099077 | enables histone-dependent DNA binding | 1 | TAS | Homo_sapiens(9606) | Function |
| GO:0099609 | enables microtubule lateral binding | 1 | IMP | Homo_sapiens(9606) | Function |
| GO:1900034 | involved_in regulation of cellular response to heat | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:1900452 | involved_in regulation of long-term synaptic depression | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:1901216 | involved_in positive regulation of neuron death | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:1902474 | involved_in positive regulation of protein localization to synapse | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:1902936 | enables phosphatidylinositol bisphosphate binding | 1 | TAS | Homo_sapiens(9606) | Function |
| GO:1902988 | involved_in neurofibrillary tangle assembly | 1 | NAS | Homo_sapiens(9606) | Process |
| GO:1903748 | involved_in negative regulation of establishment of protein localization to mitochondrion | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:1903829 | involved_in positive regulation of protein localization | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:1904115 | located_in axon cytoplasm | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:1904428 | involved_in negative regulation of tubulin deacetylation | 2 | IGI,TAS | Homo_sapiens(9606) | Process |
| GO:1905689 | involved_in positive regulation of diacylglycerol kinase activity | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:1990000 | involved_in amyloid fibril formation | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:1990090 | involved_in cellular response to nerve growth factor stimulus | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:1990416 | involved_in cellular response to brain-derived neurotrophic factor stimulus | 1 | TAS | Homo_sapiens(9606) | Process |