Welcome to
RBP World!
Description
| Ensembl ID | ENSG00000181019 | Gene ID | 1728 | Accession | 2874 |
| Symbol | NQO1 | Alias | DTD;QR1;DHQU;DIA4;NMOR1;NMORI | Full Name | NAD(P)H quinone dehydrogenase 1 |
| Status | Confidence | Length | 19673 bases | Strand | Minus strand |
| Position | 16 : 69706996 - 69726668 | RNA binding domain | N.A. | ||
| Summary | This gene is a member of the NAD(P)H dehydrogenase (quinone) family and encodes a cytoplasmic 2-electron reductase. This FAD-binding protein forms homodimers and reduces quinones to hydroquinones. This protein's enzymatic activity prevents the one electron reduction of quinones that results in the production of radical species. Mutations in this gene have been associated with tardive dyskinesia (TD), an increased risk of hematotoxicity after exposure to benzene, and susceptibility to various forms of cancer. Altered expression of this protein has been seen in many tumors and is also associated with Alzheimer's disease (AD). Alternate transcriptional splice variants, encoding different isoforms, have been characterized. [provided by RefSeq, Jul 2008] | ||||
RNA binding domains (RBDs)
| Protein | Domain | Pfam ID | E-value | Domain number |
|---|
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|---|---|---|---|
| 27515817 | Novel RNA-binding activity of NQO1 promotes SERPINA1 mRNA translation. | Andrea A Di | 2016-10-01 | Free radical biology & medicine |
| 33360352 | A redox-mediated conformational change in NQO1 controls binding to microtubules and α-tubulin acetylation. | David Siegel | 2021-02-01 | Redox biology |
| 37002649 | NQO1 regulates expression and alternative splicing of apoptotic genes associated with Alzheimer's disease in PC12 cells. | Yingshi Du | 2023-05-01 | Brain and behavior |
| 33153185 | Naturally-Occurring Rare Mutations Cause Mild to Catastrophic Effects in the Multifunctional and Cancer-Associated NQO1 Protein. | Luis Juan Pacheco-García | 2020-11-03 | Journal of personalized medicine |
| 31470592 | Nutritional Stress in Head and Neck Cancer Originating Cell Lines: The Sensitivity of the NRF2-NQO1 Axis. | Lidija Milković | 2019-08-29 | Cells |
| 28883796 | Functions of NQO1 in Cellular Protection and CoQ10 Metabolism and its Potential Role as a Redox Sensitive Molecular Switch. | David Ross | 2017-01-01 | Frontiers in physiology |
| 36349191 | Serpin family A member 1 is an oncogene in glioma and its translation is enhanced by NAD(P)H quinone dehydrogenase 1 through RNA-binding activity. | Wenjun Liu | 2022-01-01 | Open medicine (Warsaw, Poland) |
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| NQO1-205 | ENST00000561500 | 891 | 269aa | ENSP00000456282 |
| NQO1-201 | ENST00000320623 | 2521 | 274aa | ENSP00000319788 |
| NQO1-203 | ENST00000379047 | 2527 | 240aa | ENSP00000368335 |
| NQO1-202 | ENST00000379046 | 1105 | 236aa | ENSP00000368334 |
| NQO1-204 | ENST00000439109 | 924 | 202aa | ENSP00000398330 |
| NQO1-206 | ENST00000564043 | 892 | 253aa | ENSP00000455020 |
| NQO1-207 | ENST00000569118 | 519 | No protein | - |
Phenotypes
| ensgID | Trait | pValue | Pubmed ID |
|---|
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
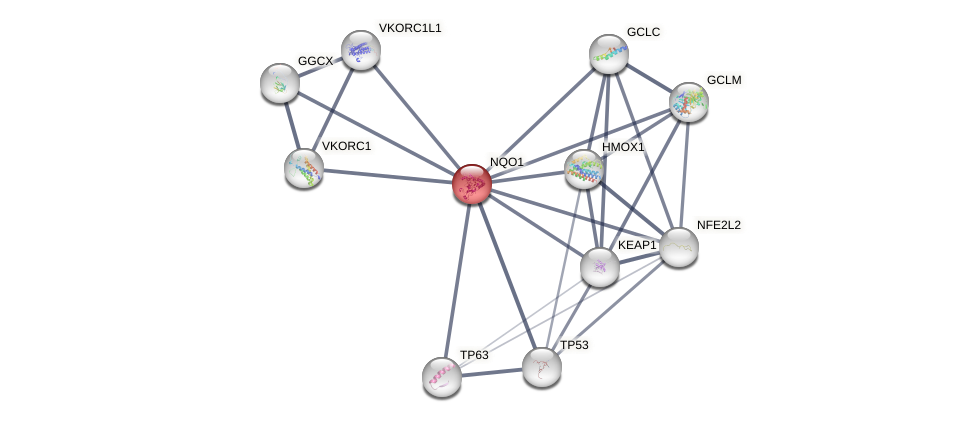
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000181019 | homo_sapiens | ENSP00000369822 | 61.4719 | 47.1861 | homo_sapiens | ENSP00000319788 | 51.8248 | 39.781 |
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 93.4307 | 92.3358 | nomascus_leucogenys | ENSNLEP00000040121 | 98.0843 | 96.9349 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 84.6715 | 84.6715 | pan_paniscus | ENSPPAP00000020607 | 83.7545 | 83.7545 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 99.2701 | 98.9051 | pongo_abelii | ENSPPYP00000034498 | 99.2701 | 98.9051 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 100 | 100 | pan_troglodytes | ENSPTRP00000014148 | 100 | 100 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 100 | 100 | gorilla_gorilla | ENSGGOP00000003236 | 100 | 100 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 92.7007 | 85.0365 | ornithorhynchus_anatinus | ENSOANP00000037586 | 92.7007 | 85.0365 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 89.781 | 79.927 | sarcophilus_harrisii | ENSSHAP00000017933 | 89.781 | 79.927 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 74.0876 | 66.0584 | notamacropus_eugenii | ENSMEUP00000014571 | 75.1852 | 67.037 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 93.7956 | 87.9562 | choloepus_hoffmanni | ENSCHOP00000011877 | 95.1852 | 89.2593 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 94.8905 | 91.2409 | dasypus_novemcinctus | ENSDNOP00000034701 | 94.8905 | 91.2409 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 71.5328 | 66.0584 | erinaceus_europaeus | ENSEEUP00000001927 | 91.1628 | 84.186 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 93.0657 | 86.4964 | echinops_telfairi | ENSETEP00000012319 | 93.4066 | 86.8132 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 95.9854 | 94.8905 | callithrix_jacchus | ENSCJAP00000073807 | 93.9286 | 92.8571 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 98.1752 | 96.7153 | cercocebus_atys | ENSCATP00000008583 | 98.1752 | 96.7153 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 97.8102 | 96.3504 | macaca_fascicularis | ENSMFAP00000005532 | 97.8102 | 96.3504 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 97.8102 | 96.7153 | macaca_mulatta | ENSMMUP00000081006 | 97.8102 | 96.7153 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 97.8102 | 96.7153 | macaca_nemestrina | ENSMNEP00000018731 | 97.8102 | 96.7153 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 97.8102 | 96.3504 | papio_anubis | ENSPANP00000029517 | 98.1685 | 96.7033 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 98.1752 | 96.7153 | mandrillus_leucophaeus | ENSMLEP00000029825 | 98.1752 | 96.7153 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 66.7883 | 63.1387 | tursiops_truncatus | ENSTTRP00000009501 | 92.4242 | 87.3737 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 38.6861 | 37.2263 | vulpes_vulpes | ENSVVUP00000023575 | 97.2477 | 93.578 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 91.6058 | 86.4964 | mus_spretus | MGP_SPRETEiJ_P0089449 | 91.6058 | 86.4964 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 92.3358 | 83.2117 | equus_asinus | ENSEASP00005055352 | 92.3358 | 83.2117 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 93.7956 | 90.146 | capra_hircus | ENSCHIP00000031320 | 93.7956 | 90.146 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 93.0657 | 89.4161 | loxodonta_africana | ENSLAFP00000007532 | 93.4066 | 89.7436 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 94.1606 | 90.146 | panthera_leo | ENSPLOP00000029703 | 94.1606 | 90.146 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 95.2555 | 91.6058 | ailuropoda_melanoleuca | ENSAMEP00000007093 | 95.6044 | 91.9414 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 94.1606 | 90.8759 | bos_indicus_hybrid | ENSBIXP00005022867 | 94.1606 | 90.8759 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 93.0657 | 88.3212 | oryctolagus_cuniculus | ENSOCUP00000026851 | 93.0657 | 88.3212 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 92.3358 | 83.2117 | equus_caballus | ENSECAP00000073897 | 86.3481 | 77.8157 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 93.0657 | 87.2263 | canis_lupus_familiaris | ENSCAFP00845012540 | 93.4066 | 87.5458 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 94.1606 | 90.146 | panthera_pardus | ENSPPRP00000025630 | 94.1606 | 90.146 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 92.3358 | 88.3212 | marmota_marmota_marmota | ENSMMMP00000015767 | 92.3358 | 88.3212 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 67.1533 | 62.4088 | balaenoptera_musculus | ENSBMSP00010020210 | 86.385 | 80.2817 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 90.5109 | 84.3066 | cavia_porcellus | ENSCPOP00000011185 | 90.1818 | 84 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 94.5256 | 89.781 | felis_catus | ENSFCAP00000018197 | 94.5256 | 89.781 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 90.146 | 86.8613 | ursus_americanus | ENSUAMP00000037174 | 94.636 | 91.1877 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 88.6861 | 81.0219 | monodelphis_domestica | ENSMODP00000003283 | 88.6861 | 81.0219 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 94.1606 | 90.8759 | bison_bison_bison | ENSBBBP00000008723 | 94.1606 | 90.8759 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 66.4234 | 63.1387 | monodon_monoceros | ENSMMNP00015004638 | 91.9192 | 87.3737 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 94.8905 | 89.4161 | sus_scrofa | ENSSSCP00000063923 | 93.8628 | 88.4477 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 94.5256 | 90.8759 | microcebus_murinus | ENSMICP00000011020 | 94.5256 | 90.8759 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 90.5109 | 83.5766 | octodon_degus | ENSODEP00000020731 | 90.8425 | 83.8828 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 88.3212 | 81.3869 | octodon_degus | ENSODEP00000019910 | 88.9706 | 81.9853 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 90.5109 | 84.6715 | jaculus_jaculus | ENSJJAP00000014502 | 91.5129 | 85.6089 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 94.1606 | 90.5109 | ovis_aries_rambouillet | ENSOARP00020012800 | 94.1606 | 90.5109 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 88.3212 | 86.1314 | ursus_maritimus | ENSUMAP00000025585 | 95.6522 | 93.2806 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 40.8759 | 37.2263 | ochotona_princeps | ENSOPRP00000009737 | 66.2722 | 60.355 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 49.2701 | 42.7007 | delphinapterus_leucas | ENSDLEP00000002343 | 71.4286 | 61.9048 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 65.3285 | 61.6788 | physeter_catodon | ENSPCTP00005025515 | 90.8629 | 85.7868 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 91.6058 | 86.1314 | mus_caroli | MGP_CAROLIEiJ_P0085950 | 91.6058 | 86.1314 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 91.9708 | 86.4964 | mus_musculus | ENSMUSP00000003947 | 91.9708 | 86.4964 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 93.0657 | 86.1314 | dipodomys_ordii | ENSDORP00000000457 | 92.0578 | 85.1986 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 94.8905 | 91.2409 | mustela_putorius_furo | ENSMPUP00000007374 | 65.8228 | 63.2911 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 70.073 | 69.3431 | aotus_nancymaae | ENSANAP00000022397 | 75.2941 | 74.5098 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 83.5766 | 82.4818 | saimiri_boliviensis_boliviensis | ENSSBOP00000020389 | 91.6 | 90.4 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 95.2555 | 92.3358 | vicugna_pacos | ENSVPAP00000001882 | 95.2555 | 92.3358 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 88.3212 | 85.4015 | chlorocebus_sabaeus | ENSCSAP00000000511 | 94.902 | 91.7647 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 91.6058 | 87.2263 | tupaia_belangeri | ENSTBEP00000012585 | 92.963 | 88.5185 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 90.8759 | 85.7664 | mesocricetus_auratus | ENSMAUP00000006795 | 90.5455 | 85.4545 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 59.4891 | 52.9197 | phocoena_sinus | ENSPSNP00000017320 | 63.1783 | 56.2015 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 94.1606 | 91.2409 | camelus_dromedarius | ENSCDRP00005027867 | 94.1606 | 91.2409 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 94.5256 | 91.6058 | carlito_syrichta | ENSTSYP00000031886 | 94.5256 | 91.6058 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 94.1606 | 90.146 | panthera_tigris_altaica | ENSPTIP00000010314 | 94.1606 | 90.146 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 91.2409 | 85.4015 | rattus_norvegicus | ENSRNOP00000082814 | 91.2409 | 85.4015 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 95.2555 | 90.8759 | prolemur_simus | ENSPSMP00000018339 | 83.3866 | 79.5527 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 87.9562 | 78.1022 | microtus_ochrogaster | ENSMOCP00000022608 | 87.9562 | 78.1022 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 87.2263 | 78.1022 | microtus_ochrogaster | ENSMOCP00000009226 | 87.2263 | 78.1022 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 80.292 | 76.2774 | sorex_araneus | ENSSARP00000010760 | 81.1808 | 77.1218 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 91.2409 | 87.5912 | peromyscus_maniculatus_bairdii | ENSPEMP00000016744 | 91.2409 | 87.5912 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 94.5256 | 91.2409 | bos_mutus | ENSBMUP00000001531 | 94.5256 | 91.2409 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 91.6058 | 86.1314 | mus_spicilegus | ENSMSIP00000003877 | 91.6058 | 86.1314 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 90.8759 | 86.1314 | cricetulus_griseus_chok1gshd | ENSCGRP00001019494 | 90.8759 | 86.1314 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 91.6058 | 83.9416 | pteropus_vampyrus | ENSPVAP00000012568 | 92.963 | 85.1852 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 94.8905 | 89.4161 | myotis_lucifugus | ENSMLUP00000012881 | 94.5455 | 89.0909 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 89.0511 | 81.3869 | vombatus_ursinus | ENSVURP00010026428 | 89.0511 | 81.3869 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 93.0657 | 88.6861 | rhinolophus_ferrumequinum | ENSRFEP00010010532 | 92.7273 | 88.3636 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 95.2555 | 92.3358 | neovison_vison | ENSNVIP00000009736 | 95.2555 | 92.3358 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 93.7956 | 90.5109 | bos_taurus | ENSBTAP00000071181 | 92.446 | 89.2086 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 85.4015 | 81.7518 | heterocephalus_glaber_female | ENSHGLP00000031476 | 92.4901 | 88.5376 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 61.6788 | 58.0292 | heterocephalus_glaber_female | ENSHGLP00000008260 | 89.8936 | 84.5745 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 98.9051 | 98.5401 | rhinopithecus_bieti | ENSRBIP00000022287 | 98.9051 | 98.5401 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 86.4964 | 82.4818 | canis_lupus_dingo | ENSCAFP00020025805 | 93.6759 | 89.3281 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 93.0657 | 87.9562 | sciurus_vulgaris | ENSSVLP00005030679 | 93.0657 | 87.9562 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 91.2409 | 80.6569 | phascolarctos_cinereus | ENSPCIP00000045627 | 61.5764 | 54.4335 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 74.8175 | 68.6131 | procavia_capensis | ENSPCAP00000001078 | 74.5455 | 68.3636 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 91.6058 | 84.3066 | nannospalax_galili | ENSNGAP00000017255 | 83.1126 | 76.4901 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 94.5256 | 90.8759 | bos_grunniens | ENSBGRP00000009205 | 83.8188 | 80.5825 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 93.0657 | 87.2263 | chinchilla_lanigera | ENSCLAP00000005194 | 93.0657 | 87.2263 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 94.5256 | 91.2409 | cervus_hanglu_yarkandensis | ENSCHYP00000031690 | 94.5256 | 91.2409 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 93.7956 | 86.1314 | catagonus_wagneri | ENSCWAP00000004693 | 83.9869 | 77.1242 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 92.7007 | 89.0511 | ictidomys_tridecemlineatus | ENSSTOP00000014369 | 92.3636 | 88.7273 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 59.854 | 58.0292 | otolemur_garnettii | ENSOGAP00000004095 | 96.4706 | 93.5294 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 96.3504 | 94.1606 | cebus_imitator | ENSCCAP00000032148 | 80.4878 | 78.6585 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 93.0657 | 89.4161 | urocitellus_parryii | ENSUPAP00010030266 | 93.0657 | 89.4161 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 94.1606 | 90.8759 | moschus_moschiferus | ENSMMSP00000004468 | 94.1606 | 90.8759 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 98.9051 | 98.5401 | rhinopithecus_roxellana | ENSRROP00000041970 | 98.9051 | 98.5401 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 92.7007 | 86.8613 | mus_pahari | MGP_PahariEiJ_P0054203 | 92.7007 | 86.8613 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 94.1606 | 91.6058 | propithecus_coquereli | ENSPCOP00000018768 | 95.5556 | 92.963 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 69.708 | 55.1095 | latimeria_chalumnae | ENSLACP00000009170 | 75.1969 | 59.4488 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 68.9781 | 50 | lepisosteus_oculatus | ENSLOCP00000003285 | 64.9485 | 47.079 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 71.1679 | 49.635 | danio_rerio | ENSDARP00000021150 | 70.9091 | 49.4545 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 68.9781 | 49.635 | carassius_auratus | ENSCARP00000081609 | 68.4783 | 49.2754 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 68.6131 | 53.2847 | astyanax_mexicanus | ENSAMXP00000036782 | 68.1159 | 52.8986 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 65.3285 | 50.7299 | astyanax_mexicanus | ENSAMXP00000032046 | 67.5472 | 52.4528 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 70.438 | 53.6496 | ictalurus_punctatus | ENSIPUP00000016830 | 70.696 | 53.8462 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 68.2482 | 49.635 | esox_lucius | ENSELUP00000016459 | 62.1262 | 45.1827 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 67.8832 | 50 | esox_lucius | ENSELUP00000067439 | 68.8889 | 50.7407 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 67.8832 | 50 | esox_lucius | ENSELUP00000075929 | 68.3824 | 50.3676 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 68.9781 | 50.7299 | esox_lucius | ENSELUP00000072616 | 70 | 51.4815 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 68.6131 | 51.0949 | esox_lucius | ENSELUP00000073421 | 69.6296 | 51.8519 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 68.9781 | 51.4599 | esox_lucius | ENSELUP00000065434 | 69.4853 | 51.8382 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 68.2482 | 49.635 | esox_lucius | ENSELUP00000028520 | 68.4982 | 49.8168 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 68.2482 | 50 | esox_lucius | ENSELUP00000052361 | 66.3121 | 48.5816 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 37.9562 | 27.3723 | esox_lucius | ENSELUP00000061315 | 69.3333 | 50 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 72.2628 | 51.0949 | oncorhynchus_kisutch | ENSOKIP00005047642 | 70.4626 | 49.8221 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 72.2628 | 51.0949 | oncorhynchus_kisutch | ENSOKIP00005071363 | 70.4626 | 49.8221 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 71.1679 | 51.0949 | oncorhynchus_mykiss | ENSOMYP00000057873 | 70.3971 | 50.5415 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 71.1679 | 51.0949 | oncorhynchus_mykiss | ENSOMYP00000051620 | 70.3971 | 50.5415 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 72.2628 | 51.0949 | salmo_salar | ENSSSAP00000021597 | 68.0412 | 48.11 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 67.5182 | 51.4599 | salmo_salar | ENSSSAP00000058088 | 59.8706 | 45.6311 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 71.1679 | 51.0949 | salmo_salar | ENSSSAP00000165983 | 67.474 | 48.4429 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 71.8978 | 52.1898 | salmo_trutta | ENSSTUP00000077536 | 71.8978 | 52.1898 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 67.8832 | 51.0949 | salmo_trutta | ENSSTUP00000018073 | 68.3824 | 51.4706 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 72.2628 | 52.1898 | salmo_trutta | ENSSTUP00000002334 | 72.2628 | 52.1898 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 42.7007 | 30.6569 | gadus_morhua | ENSGMOP00000004034 | 69.2308 | 49.7041 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 63.5037 | 48.5401 | fundulus_heteroclitus | ENSFHEP00000027673 | 63.2727 | 48.3636 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 58.0292 | 41.9708 | poecilia_reticulata | ENSPREP00000013769 | 62.3529 | 45.098 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 68.9781 | 53.2847 | xiphophorus_maculatus | ENSXMAP00000021292 | 67.5 | 52.1429 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 55.1095 | 42.3358 | oryzias_latipes | ENSORLP00000022373 | 65.6522 | 50.4348 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 32.8467 | 22.9927 | oryzias_latipes | ENSORLP00000022376 | 66.1765 | 46.3235 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 66.7883 | 49.2701 | cyclopterus_lumpus | ENSCLMP00005031573 | 65.3571 | 48.2143 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 66.0584 | 48.5401 | cyclopterus_lumpus | ENSCLMP00005031582 | 64.6429 | 47.5 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 35.0365 | 24.4526 | astatotilapia_calliptera | ENSACLP00000002288 | 68.5714 | 47.8571 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 35.0365 | 24.4526 | astatotilapia_calliptera | ENSACLP00000002305 | 66.2069 | 46.2069 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 75.5474 | 62.7737 | xenopus_tropicalis | ENSXETP00000093423 | 72.3776 | 60.1399 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 80.292 | 66.0584 | chrysemys_picta_bellii | ENSCPBP00000039251 | 74.0741 | 60.9428 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 80.6569 | 64.5985 | sphenodon_punctatus | ENSSPUP00000007448 | 82.7715 | 66.2921 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 85.0365 | 69.708 | crocodylus_porosus | ENSCPRP00005011913 | 70.8207 | 58.0547 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 67.5182 | 54.3796 | notechis_scutatus | ENSNSUP00000028665 | 57.4534 | 46.2733 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 76.6423 | 63.1387 | pseudonaja_textilis | ENSPTXP00000011983 | 57.6923 | 47.5275 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 76.6423 | 59.854 | anas_platyrhynchos_platyrhynchos | ENSAPLP00000016678 | 60.6936 | 47.3988 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 72.9927 | 55.8394 | gallus_gallus | ENSGALP00010011356 | 77.821 | 59.5331 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 78.4672 | 59.854 | meleagris_gallopavo | ENSMGAP00000008896 | 55.5556 | 42.3773 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 79.927 | 64.9635 | anolis_carolinensis | ENSACAP00000003582 | 80.2198 | 65.2015 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 55.8394 | 41.6058 | neolamprologus_brichardi | ENSNBRP00000014146 | 52.3973 | 39.0411 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 84.6715 | 70.073 | podarcis_muralis | ENSPMRP00000012865 | 84.3636 | 69.8182 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 71.8978 | 52.9197 | erpetoichthys_calabaricus | ENSECRP00000028479 | 72.1612 | 53.1136 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 54.3796 | 36.4963 | kryptolebias_marmoratus | ENSKMAP00000024709 | 61.0656 | 40.9836 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 82.1168 | 65.6934 | salvator_merianae | ENSSMRP00000009136 | 81.5217 | 65.2174 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 56.5693 | 40.8759 | amphilophus_citrinellus | ENSACIP00000025301 | 63.2653 | 45.7143 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 69.708 | 52.5547 | amphiprion_percula | ENSAPEP00000000080 | 69.2029 | 52.1739 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 73.7226 | 56.9343 | pelodiscus_sinensis | ENSPSIP00000013634 | 79.5276 | 61.4173 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 79.1971 | 66.0584 | gopherus_evgoodei | ENSGEVP00005019828 | 78.9091 | 65.8182 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 71.1679 | 59.4891 | chelonoidis_abingdonii | ENSCABP00000010876 | 78.9474 | 65.9919 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 66.0584 | 49.2701 | seriola_dumerili | ENSSDUP00000005493 | 66.5441 | 49.6324 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 66.7883 | 50 | cottoperca_gobio | ENSCGOP00000047895 | 67.2794 | 50.3676 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 57.2993 | 44.1606 | anser_brachyrhynchus | ENSABRP00000002981 | 28.5455 | 22 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 67.8832 | 48.9051 | gasterosteus_aculeatus | ENSGACP00000004955 | 68.3824 | 49.2647 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 56.2044 | 40.146 | gasterosteus_aculeatus | ENSGACP00000004918 | 70.3196 | 50.2283 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 67.8832 | 48.9051 | gasterosteus_aculeatus | ENSGACP00000004936 | 66.9065 | 48.2014 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 62.7737 | 44.8905 | gasterosteus_aculeatus | ENSGACP00000004940 | 68.8 | 49.2 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 45.9854 | 35.7664 | poecilia_formosa | ENSPFOP00000015639 | 68.4783 | 53.2609 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 67.5182 | 48.1752 | sander_lucioperca | ENSSLUP00000001832 | 61.6667 | 44 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 64.2336 | 46.3504 | sander_lucioperca | ENSSLUP00000019442 | 67.6923 | 48.8462 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 70.073 | 53.2847 | amphiprion_ocellaris | ENSAOCP00000008190 | 69.5652 | 52.8986 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 79.562 | 64.5985 | terrapene_carolina_triunguis | ENSTMTP00000027637 | 79.2727 | 64.3636 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 79.562 | 64.5985 | terrapene_carolina_triunguis | ENSTMTP00000026157 | 79.2727 | 64.3636 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 64.2336 | 48.9051 | hucho_hucho | ENSHHUP00000055732 | 69.5652 | 52.9644 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 46.7153 | 32.8467 | hucho_hucho | ENSHHUP00000005334 | 66.6667 | 46.875 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 36.1314 | 27.0073 | hucho_hucho | ENSHHUP00000011320 | 70.2128 | 52.4823 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 71.8978 | 50 | betta_splendens | ENSBSLP00000020235 | 69.3662 | 48.2394 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 67.5182 | 51.4599 | pygocentrus_nattereri | ENSPNAP00000022807 | 66.5468 | 50.7194 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 66.0584 | 49.2701 | seriola_lalandi_dorsalis | ENSSLDP00000026520 | 66.5441 | 49.6324 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 66.0584 | 47.4453 | labrus_bergylta | ENSLBEP00000021291 | 65.8182 | 47.2727 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 62.7737 | 47.0803 | mastacembelus_armatus | ENSMAMP00000021102 | 67.7165 | 50.7874 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 33.5766 | 23.3577 | stegastes_partitus | ENSSPAP00000009637 | 68.6567 | 47.7612 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 63.1387 | 44.8905 | oncorhynchus_tshawytscha | ENSOTSP00005049956 | 60.4895 | 43.007 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 37.2263 | 28.4672 | oncorhynchus_tshawytscha | ENSOTSP00005079946 | 58.6207 | 44.8276 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 70.438 | 53.2847 | nothobranchius_furzeri | ENSNFUP00015006418 | 67.2474 | 50.8711 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 54.3796 | 40.146 | nothobranchius_furzeri | ENSNFUP00015025806 | 53.7906 | 39.7112 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 66.0584 | 48.9051 | denticeps_clupeoides | ENSDCDP00000027183 | 67.7903 | 50.1873 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 66.4234 | 48.9051 | denticeps_clupeoides | ENSDCDP00000026064 | 68.4211 | 50.3759 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 66.0584 | 48.9051 | denticeps_clupeoides | ENSDCDP00000026099 | 67.7903 | 50.1873 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 65.6934 | 48.9051 | larimichthys_crocea | ENSLCRP00005028642 | 66.1765 | 49.2647 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 65.3285 | 46.7153 | larimichthys_crocea | ENSLCRP00005005964 | 66.2963 | 47.4074 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 70.073 | 50.7299 | cyprinus_carpio_carpio | ENSCCRP00000129509 | 68.0851 | 49.2908 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 79.1971 | 60.9489 | coturnix_japonica | ENSCJPP00005018750 | 63.0814 | 48.5465 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 68.9781 | 53.2847 | poecilia_latipinna | ENSPLAP00000022156 | 67.5 | 52.1429 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 35.7664 | 25.5474 | sinocyclocheilus_grahami | ENSSGRP00000013467 | 58.3333 | 41.6667 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 61.3139 | 45.6204 | maylandia_zebra | ENSMZEP00005003590 | 66.9323 | 49.8008 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 56.9343 | 39.781 | maylandia_zebra | ENSMZEP00005003458 | 63.9344 | 44.6721 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 60.9489 | 44.8905 | maylandia_zebra | ENSMZEP00005003492 | 66.5339 | 49.004 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 69.3431 | 54.3796 | scleropages_formosus | ENSSFOP00015030056 | 67.6157 | 53.0249 |
| ENSG00000181019 | homo_sapiens | ENSP00000319788 | 75.9124 | 59.854 | leptobrachium_leishanense | ENSLLEP00000042760 | 66.4537 | 52.3962 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0000209 | involved_in protein polyubiquitination | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0002931 | involved_in response to ischemia | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0003723 | enables RNA binding | 1 | HDA | Homo_sapiens(9606) | Function |
| GO:0003955 | enables NAD(P)H dehydrogenase (quinone) activity | 1 | IBA | Homo_sapiens(9606) | Function |
| GO:0004128 | enables cytochrome-b5 reductase activity, acting on NAD(P)H | 1 | TAS | Homo_sapiens(9606) | Function |
| GO:0004784 | enables superoxide dismutase activity | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005634 | located_in nucleus | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0005737 | located_in cytoplasm | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0005829 | is_active_in cytosol | 3 | IBA,IDA,TAS | Homo_sapiens(9606) | Component |
| GO:0006116 | involved_in NADH oxidation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0006743 | involved_in ubiquinone metabolic process | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0006805 | involved_in xenobiotic metabolic process | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0006809 | involved_in nitric oxide biosynthetic process | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0006979 | involved_in response to oxidative stress | 1 | IEP | Homo_sapiens(9606) | Process |
| GO:0007271 | involved_in synaptic transmission, cholinergic | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0007584 | involved_in response to nutrient | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0008753 | enables NADPH dehydrogenase (quinone) activity | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0009636 | involved_in response to toxic substance | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0009725 | involved_in response to hormone | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0009743 | involved_in response to carbohydrate | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0014075 | involved_in response to amine | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0019430 | involved_in removal of superoxide radicals | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0030163 | involved_in protein catabolic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0030425 | located_in dendrite | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0032355 | involved_in response to estradiol | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0032496 | involved_in response to lipopolysaccharide | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0033574 | involved_in response to testosterone | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0034599 | involved_in cellular response to oxidative stress | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0042177 | involved_in negative regulation of protein catabolic process | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0042360 | involved_in vitamin E metabolic process | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0042373 | involved_in vitamin K metabolic process | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0042802 | enables identical protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0043025 | located_in neuronal cell body | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0043066 | involved_in negative regulation of apoptotic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0043086 | involved_in negative regulation of catalytic activity | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0043279 | involved_in response to alkaloid | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0043525 | involved_in positive regulation of neuron apoptotic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0045087 | involved_in innate immune response | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0045202 | located_in synapse | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0045454 | involved_in cell redox homeostasis | 1 | IEP | Homo_sapiens(9606) | Process |
| GO:0045471 | involved_in response to ethanol | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0050136 | enables NADH dehydrogenase (quinone) activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0051602 | involved_in response to electrical stimulus | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0070301 | involved_in cellular response to hydrogen peroxide | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0070995 | involved_in NADPH oxidation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0071248 | involved_in cellular response to metal ion | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:1904772 | involved_in response to tetrachloromethane | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:1904844 | involved_in response to L-glutamine | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:1904880 | involved_in response to hydrogen sulfide | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:1905395 | involved_in response to flavonoid | 1 | IEA | Homo_sapiens(9606) | Process |