RBP Type
Canonical_RBPs
Diseases
SARS-COV-2
Drug
N.A.
Main interacting RNAs
N.A.
Moonlighting functions
N.A.
Localizations
Stress granucle
BulkPerturb-seq
N.A.
Ensembl ID ENSG00000179750 Gene ID
9582 Accession
17352
Symbol
APOBEC3B
Alias
A3B;ARP4;ARCD3;PHRBNL;APOBEC1L;bK150C2.2;DJ742C19.2
Full Name
apolipoprotein B mRNA editing enzyme catalytic subunit 3B
Status
Confidence
Length
10458 bases
Strand
Plus strand
Position
22 : 38982347 - 38992804
RNA binding domain
APOBEC_N , APOBEC_C
Summary
This gene is a member of the cytidine deaminase gene family. It is one of seven related genes or pseudogenes found in a cluster, thought to result from gene duplication, on chromosome 22. Members of the cluster encode proteins that are structurally and functionally related to the C to U RNA-editing cytidine deaminase APOBEC1. It is thought that the proteins may be RNA editing enzymes and have roles in growth or cell cycle control. A hybrid gene results from the deletion of approximately 29.5 kb of sequence between this gene, APOBEC3B, and the adjacent gene APOBEC3A. The breakpoints of the deletion are within the two genes, so the deletion allele is predicted to have the promoter and coding region of APOBEC3A, but the 3' UTR of APOBEC3B. Two transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jul 2012]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
ENSP00000327459
APOBEC_N
PF08210 4.2e-100
1
ENSP00000327459
APOBEC_N
PF08210 4.2e-100
2
ENSP00000327459
APOBEC_C
PF05240 2.4e-62
3
ENSP00000327459
APOBEC_C
PF05240 2.4e-62
4
ENSP00000385060
APOBEC_N
PF08210 1.2e-99
1
ENSP00000385060
APOBEC_N
PF08210 1.2e-99
2
ENSP00000385060
APOBEC_C
PF05240 5.7e-62
3
ENSP00000385060
APOBEC_C
PF05240 5.7e-62
4
ENSP00000385068
APOBEC_N
PF08210 3.5e-91
1
ENSP00000385068
APOBEC_N
PF08210 3.5e-91
2
ENSP00000385068
APOBEC_C
PF05240 1.9e-62
3
ENSP00000385068
APOBEC_C
PF05240 1.9e-62
4
ENSP00000338897
APOBEC_N
PF08210 2.3e-47
1
ENSP00000338897
APOBEC_C
PF05240 6e-32
2
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
36978112 Host-mediated RNA editing in viruses.
Tongtong Zhu
2023-03-28
Biology direct
33661412 Identification of anti-Epstein-Barr virus (EBV) antibody signature in EBV-associated gastric carcinoma.
Lusheng Song
2021-07-01
Gastric cancer : official journal of the International Gastric Cancer Association and the Japanese Gastric Cancer Association
30679582 A panel of eGFP reporters for single base editing by APOBEC-Cas9 editosome complexes.
St A Martin
2019-01-24
Scientific reports
Name
Transcript ID
bp
Protein
Translation ID
APOBEC3B-204
ENST00000407298
1282
357aa
ENSP00000385068
APOBEC3B-201
ENST00000333467
1590
382aa
ENSP00000327459
APOBEC3B-203
ENST00000402182
1844
490aa
ENSP00000385060
APOBEC3B-202
ENST00000335760
1359
251aa
ENSP00000338897
Pathway ID
Pathway Name
Source
ensgID
Trait
pValue
Pubmed ID
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
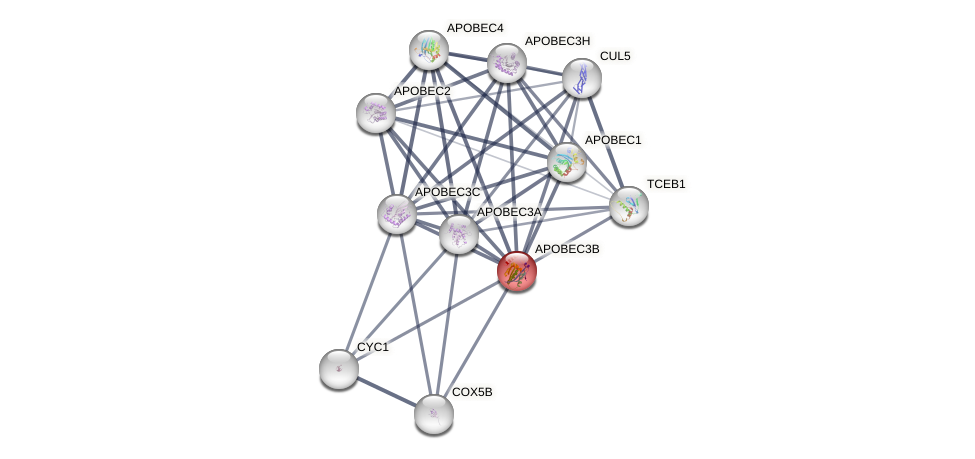
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
ENSG00000179750 homo_sapiens ENSP00000229335 53.0303 39.3939 homo_sapiens ENSP00000327459 27.4869 20.4188 ENSG00000179750 homo_sapiens ENSP00000385057 69.0104 56.25 homo_sapiens ENSP00000327459 69.3717 56.5445 ENSG00000179750 homo_sapiens ENSP00000411754 54.6448 31.1475 homo_sapiens ENSP00000327459 26.178 14.9215 ENSG00000179750 homo_sapiens ENSP00000309749 69.7051 60.0536 homo_sapiens ENSP00000327459 68.0628 58.6387 ENSG00000179750 homo_sapiens ENSP00000216099 68.1347 59.5855 homo_sapiens ENSP00000327459 68.8482 60.2094 ENSG00000179750 homo_sapiens ENSP00000355340 54.7368 42.1053 homo_sapiens ENSP00000327459 27.2251 20.9424 ENSG00000179750 homo_sapiens ENSP00000229304 37.7119 22.8814 homo_sapiens ENSP00000327459 23.2984 14.1361 ENSG00000179750 homo_sapiens ENSP00000244669 47.7679 29.9107 homo_sapiens ENSP00000327459 28.0105 17.5393 ENSG00000179750 homo_sapiens ENSP00000249116 90.4523 86.9347 homo_sapiens ENSP00000327459 47.1204 45.288
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
ENSG00000179750 homo_sapiens ENSP00000327459 90.0524 84.2932 nomascus_leucogenys ENSNLEP00000018464 70.7819 66.2551 ENSG00000179750 homo_sapiens ENSP00000327459 96.3351 92.4084 pan_paniscus ENSPPAP00000002122 75.102 72.0408 ENSG00000179750 homo_sapiens ENSP00000327459 90.0524 84.8168 pongo_abelii ENSPPYP00000044599 70.2041 66.1225 ENSG00000179750 homo_sapiens ENSP00000327459 85.3403 81.6754 pan_troglodytes ENSPTRP00000024776 66.5306 63.6735 ENSG00000179750 homo_sapiens ENSP00000327459 97.1204 92.6702 gorilla_gorilla ENSGGOP00000006577 97.1204 92.6702 ENSG00000179750 homo_sapiens ENSP00000327459 20.9424 13.089 sarcophilus_harrisii ENSSHAP00000043123 27.7778 17.3611 ENSG00000179750 homo_sapiens ENSP00000327459 74.3456 65.1832 callithrix_jacchus ENSCJAP00000088528 73.9583 64.8438 ENSG00000179750 homo_sapiens ENSP00000327459 88.2199 82.199 cercocebus_atys ENSCATP00000003628 68.7755 64.0816 ENSG00000179750 homo_sapiens ENSP00000327459 44.5026 40.8377 macaca_mulatta ENSMMUP00000014095 49.7076 45.614 ENSG00000179750 homo_sapiens ENSP00000327459 89.5288 84.2932 macaca_mulatta ENSMMUP00000014096 89.5288 84.2932 ENSG00000179750 homo_sapiens ENSP00000327459 76.4398 71.2042 macaca_nemestrina ENSMNEP00000040658 83.908 78.1609 ENSG00000179750 homo_sapiens ENSP00000327459 88.7435 83.5079 mandrillus_leucophaeus ENSMLEP00000001202 88.7435 83.5079 ENSG00000179750 homo_sapiens ENSP00000327459 21.466 13.6126 monodelphis_domestica ENSMODP00000040505 31.5385 20 ENSG00000179750 homo_sapiens ENSP00000327459 36.1257 32.7225 chlorocebus_sabaeus ENSCSAP00000014407 66.0287 59.8086 ENSG00000179750 homo_sapiens ENSP00000327459 89.0052 83.2461 chlorocebus_sabaeus ENSCSAP00000002728 69.3878 64.898 ENSG00000179750 homo_sapiens ENSP00000327459 21.2042 13.6126 vombatus_ursinus ENSVURP00010031524 27.1812 17.4497 ENSG00000179750 homo_sapiens ENSP00000327459 90.0524 84.555 rhinopithecus_bieti ENSRBIP00000038646 90.0524 84.555 ENSG00000179750 homo_sapiens ENSP00000327459 21.466 14.9215 phascolarctos_cinereus ENSPCIP00000049844 31.5385 21.9231 ENSG00000179750 homo_sapiens ENSP00000327459 73.2984 62.0419 cebus_imitator ENSCCAP00000019211 72.9167 61.7188 ENSG00000179750 homo_sapiens ENSP00000327459 90.0524 84.555 rhinopithecus_roxellana ENSRROP00000008655 90.0524 84.555 ENSG00000179750 homo_sapiens ENSP00000327459 20.4188 10.733 callorhinchus_milii ENSCMIP00000004961 37.1429 19.5238 ENSG00000179750 homo_sapiens ENSP00000327459 20.6806 12.5654 latimeria_chalumnae ENSLACP00000018020 22.9651 13.9535 ENSG00000179750 homo_sapiens ENSP00000327459 22.2513 12.5654 lepisosteus_oculatus ENSLOCP00000004258 31.1355 17.5824 ENSG00000179750 homo_sapiens ENSP00000327459 23.5602 13.3508 xenopus_tropicalis ENSXETP00000073758 40.5405 22.973 ENSG00000179750 homo_sapiens ENSP00000327459 21.9895 12.5654 anolis_carolinensis ENSACAP00000018261 31.2268 17.8439 ENSG00000179750 homo_sapiens ENSP00000327459 19.6335 9.94764 podarcis_muralis ENSPMRP00000035742 35.7143 18.0952 ENSG00000179750 homo_sapiens ENSP00000327459 21.466 12.5654 erpetoichthys_calabaricus ENSECRP00000021015 31.4176 18.3908 ENSG00000179750 homo_sapiens ENSP00000327459 20.9424 12.5654 paramormyrops_kingsleyae ENSPKIP00000029857 38.6473 23.1884 ENSG00000179750 homo_sapiens ENSP00000327459 20.4188 12.0419 scleropages_formosus ENSSFOP00015009596 39.3939 23.2323
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0000932 is_active_in P-body 1 IBA Homo_sapiens(9606) Component GO:0003723 enables RNA binding 2 HDA,IBA Homo_sapiens(9606) Function GO:0004126 enables cytidine deaminase activity 1 IBA Homo_sapiens(9606) Function GO:0005515 enables protein binding 1 IPI Homo_sapiens(9606) Function GO:0005634 is_active_in nucleus 2 IBA,IDA Homo_sapiens(9606) Component GO:0005654 located_in nucleoplasm 1 IDA Homo_sapiens(9606) Component GO:0005737 is_active_in cytoplasm 1 IBA Homo_sapiens(9606) Component GO:0008270 enables zinc ion binding 1 IEA Homo_sapiens(9606) Function GO:0009972 involved_in cytidine deamination 1 IEA Homo_sapiens(9606) Process GO:0010526 involved_in retrotransposon silencing 1 IDA Homo_sapiens(9606) Process GO:0016554 involved_in cytidine to uridine editing 1 IBA Homo_sapiens(9606) Process GO:0044355 involved_in clearance of foreign intracellular DNA 1 IDA Homo_sapiens(9606) Process GO:0045087 involved_in innate immune response 1 IEA Homo_sapiens(9606) Process GO:0045869 involved_in negative regulation of single stranded viral RNA replication via double stranded DNA intermediate 1 IBA Homo_sapiens(9606) Process GO:0047844 enables deoxycytidine deaminase activity 2 IBA,IMP Homo_sapiens(9606) Function GO:0051607 involved_in defense response to virus 2 IBA,IDA Homo_sapiens(9606) Process GO:0070383 involved_in DNA cytosine deamination 1 IBA Homo_sapiens(9606) Process GO:0080111 involved_in DNA demethylation 1 IBA Homo_sapiens(9606) Process
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.