Welcome to
RBP World!
Functions and Diseases
| RBP Type | Canonical_RBPs |
| Diseases | Cancer |
| Drug | N.A. |
| Main interacting RNAs | mRNA |
| Moonlighting functions | N.A. |
| Localizations | Stress granucle |
| BulkPerturb-seq | DataSet_01_432 |
Description
| Ensembl ID | ENSG00000179218 | Gene ID | 811 | Accession | 1455 |
| Symbol | CALR | Alias | RO;CRT;SSA;CALR1;cC1qR;HEL-S-99n | Full Name | calreticulin |
| Status | Confidence | Length | 5881 bases | Strand | Plus strand |
| Position | 19 : 12938609 - 12944489 | RNA binding domain | Calreticulin | ||
| Summary | Calreticulin is a highly conserved chaperone protein which resides primarily in the endoplasmic reticulum, and is involved in a variety of cellular processes, among them, cell adhesion. Additionally, it functions in protein folding quality control and calcium homeostasis. Calreticulin is also found in the nucleus, suggesting that it may have a role in transcription regulation. Systemic lupus erythematosus is associated with increased autoantibody titers against calreticulin. Recurrent mutations in calreticulin have been linked to various neoplasms, including the myeloproliferative type.[provided by RefSeq, May 2020] | ||||
RNA binding domains (RBDs)
| Protein | Domain | Pfam ID | E-value | Domain number |
|---|---|---|---|---|
| ENSP00000465918 | Calreticulin | PF00262 | 1.1e-81 | 1 |
| ENSP00000465918 | Calreticulin | PF00262 | 1.1e-81 | 2 |
| ENSP00000320866 | Calreticulin | PF00262 | 3.7e-79 | 1 |
| ENSP00000320866 | Calreticulin | PF00262 | 3.7e-79 | 2 |
| ENSP00000466037 | Calreticulin | PF00262 | 3.7e-79 | 1 |
| ENSP00000466037 | Calreticulin | PF00262 | 3.7e-79 | 2 |
| ENSP00000504963 | Calreticulin | PF00262 | 1.5e-77 | 1 |
| ENSP00000504963 | Calreticulin | PF00262 | 1.5e-77 | 2 |
| ENSP00000465105 | Calreticulin | PF00262 | 4.2e-51 | 1 |
| ENSP00000465105 | Calreticulin | PF00262 | 4.2e-51 | 2 |
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| CALR-205 | ENST00000587486 | 957 | No protein | - |
| CALR-202 | ENST00000586760 | 1630 | 378aa | ENSP00000465918 |
| CALR-201 | ENST00000316448 | 1901 | 417aa | ENSP00000320866 |
| CALR-204 | ENST00000586967 | 1678 | 417aa | ENSP00000466037 |
| CALR-208 | ENST00000680816 | 1564 | 356aa | ENSP00000504963 |
| CALR-207 | ENST00000590325 | 781 | No protein | - |
| CALR-206 | ENST00000588454 | 1661 | 349aa | ENSP00000465105 |
| CALR-203 | ENST00000586803 | 667 | No protein | - |
Phenotypes
| ensgID | Trait | pValue | Pubmed ID |
|---|
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
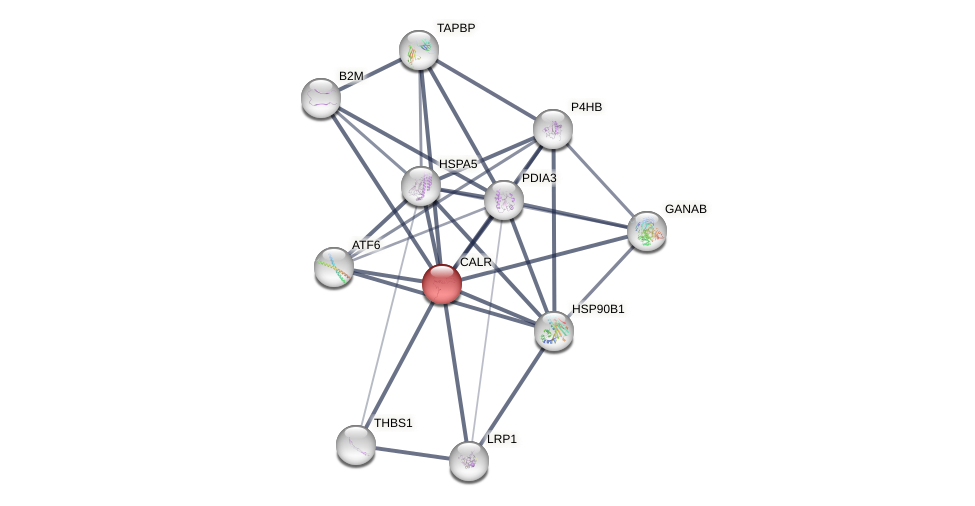
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000179218 | homo_sapiens | ENSP00000269881 | 69.7917 | 49.7396 | homo_sapiens | ENSP00000320866 | 64.2686 | 45.8034 |
| ENSG00000179218 | homo_sapiens | ENSP00000326699 | 42.459 | 28.5246 | homo_sapiens | ENSP00000320866 | 62.1103 | 41.7266 |
| ENSG00000179218 | homo_sapiens | ENSP00000247461 | 43.0743 | 30.5743 | homo_sapiens | ENSP00000320866 | 61.1511 | 43.4053 |
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 100 | 99.7602 | nomascus_leucogenys | ENSNLEP00000015731 | 100 | 99.7602 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 99.7602 | 99.5204 | pan_paniscus | ENSPPAP00000001484 | 99.7602 | 99.5204 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 100 | 100 | pongo_abelii | ENSPPYP00000010780 | 100 | 100 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 99.0408 | 99.0408 | pan_troglodytes | ENSPTRP00000018010 | 99.7585 | 99.7585 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 99.7602 | 99.5204 | gorilla_gorilla | ENSGGOP00000015732 | 99.7602 | 99.5204 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 92.0863 | 84.1727 | ornithorhynchus_anatinus | ENSOANP00000015541 | 91.6468 | 83.7709 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 85.1319 | 77.458 | sarcophilus_harrisii | ENSSHAP00000045726 | 83.9243 | 76.3593 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 81.7746 | 76.4988 | notamacropus_eugenii | ENSMEUP00000013026 | 83.5784 | 78.1863 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 75.5396 | 72.9017 | choloepus_hoffmanni | ENSCHOP00000003178 | 78.3582 | 75.6219 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 95.9233 | 93.0456 | dasypus_novemcinctus | ENSDNOP00000024904 | 95.4654 | 92.6014 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 97.1223 | 95.4436 | callithrix_jacchus | ENSCJAP00000072313 | 96.89 | 95.2153 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 83.4532 | 80.5755 | cercocebus_atys | ENSCATP00000019363 | 89.4602 | 86.3753 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 99.7602 | 99.7602 | macaca_fascicularis | ENSMFAP00000046326 | 99.7602 | 99.7602 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 99.7602 | 99.7602 | macaca_mulatta | ENSMMUP00000047382 | 99.7602 | 99.7602 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 99.7602 | 99.7602 | macaca_nemestrina | ENSMNEP00000022121 | 99.7602 | 99.7602 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 99.7602 | 99.5204 | papio_anubis | ENSPANP00000001251 | 99.7602 | 99.5204 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 99.7602 | 99.7602 | mandrillus_leucophaeus | ENSMLEP00000037878 | 99.7602 | 99.7602 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 96.4029 | 93.0456 | tursiops_truncatus | ENSTTRP00000003375 | 96.4029 | 93.0456 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 97.3621 | 94.964 | vulpes_vulpes | ENSVVUP00000033408 | 97.3621 | 94.964 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 97.1223 | 93.0456 | mus_spretus | MGP_SPRETEiJ_P0088800 | 97.3558 | 93.2692 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 95.6835 | 92.8058 | equus_asinus | ENSEASP00005062704 | 96.8447 | 93.932 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 95.9233 | 93.0456 | capra_hircus | ENSCHIP00000027670 | 95.9233 | 93.0456 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 95.4436 | 93.0456 | loxodonta_africana | ENSLAFP00000013561 | 96.1353 | 93.7198 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 96.8825 | 94.2446 | panthera_leo | ENSPLOP00000021543 | 96.8825 | 94.2446 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 91.8465 | 91.8465 | ailuropoda_melanoleuca | ENSAMEP00000035694 | 100 | 100 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 92.5659 | 89.4484 | bos_indicus_hybrid | ENSBIXP00005040423 | 92.1241 | 89.0215 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 97.3621 | 94.964 | canis_lupus_familiaris | ENSCAFP00845021381 | 97.3621 | 94.964 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 96.6427 | 94.0048 | panthera_pardus | ENSPPRP00000004410 | 96.6427 | 94.0048 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 95.9233 | 93.2854 | marmota_marmota_marmota | ENSMMMP00000023986 | 95.6938 | 93.0622 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 76.259 | 71.4628 | balaenoptera_musculus | ENSBMSP00010027637 | 88.0886 | 82.5485 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 95.6835 | 92.5659 | balaenoptera_musculus | ENSBMSP00010008310 | 95.6835 | 92.5659 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 97.1223 | 93.765 | cavia_porcellus | ENSCPOP00000000708 | 97.1223 | 93.765 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 96.8825 | 94.964 | felis_catus | ENSFCAP00000000444 | 96.8825 | 94.964 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 97.3621 | 95.4436 | ursus_americanus | ENSUAMP00000012645 | 97.3621 | 95.4436 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 91.3669 | 82.7338 | monodelphis_domestica | ENSMODP00000014423 | 91.1483 | 82.5359 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 95.9233 | 92.8058 | bison_bison_bison | ENSBBBP00000011040 | 95.9233 | 92.8058 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 96.1631 | 92.5659 | monodon_monoceros | ENSMMNP00015006511 | 96.1631 | 92.5659 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 63.789 | 58.753 | monodon_monoceros | ENSMMNP00015013294 | 61.0092 | 56.1927 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 96.6427 | 93.5252 | sus_scrofa | ENSSSCP00000014615 | 96.6427 | 93.5252 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 97.6019 | 95.2038 | microcebus_murinus | ENSMICP00000014837 | 97.6019 | 95.2038 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 97.6019 | 94.7242 | octodon_degus | ENSODEP00000014475 | 97.6019 | 94.7242 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 95.9233 | 93.2854 | jaculus_jaculus | ENSJJAP00000000098 | 97.0874 | 94.4175 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 96.1631 | 92.8058 | ovis_aries_rambouillet | ENSOARP00020012651 | 96.1631 | 92.8058 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 90.4077 | 88.0096 | ovis_aries_rambouillet | ENSOARP00020016621 | 96.1735 | 93.6225 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 97.3621 | 95.4436 | ursus_maritimus | ENSUMAP00000007936 | 97.3621 | 95.4436 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 67.3861 | 63.3094 | ochotona_princeps | ENSOPRP00000004620 | 72.4227 | 68.0412 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 95.9233 | 92.5659 | delphinapterus_leucas | ENSDLEP00000014630 | 95.9233 | 92.5659 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 96.4029 | 92.8058 | physeter_catodon | ENSPCTP00005027220 | 96.4029 | 92.8058 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 97.6019 | 93.5252 | mus_caroli | MGP_CAROLIEiJ_P0085300 | 97.8365 | 93.75 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 97.6019 | 94.0048 | mus_musculus | ENSMUSP00000003912 | 97.8365 | 94.2308 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 97.6019 | 95.4436 | dipodomys_ordii | ENSDORP00000023014 | 97.6019 | 95.4436 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 97.6019 | 95.4436 | mustela_putorius_furo | ENSMPUP00000004315 | 97.6019 | 95.4436 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 96.4029 | 95.2038 | aotus_nancymaae | ENSANAP00000006562 | 94.8113 | 93.6321 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 99.2806 | 97.8417 | saimiri_boliviensis_boliviensis | ENSSBOP00000004743 | 99.2806 | 97.8417 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 99.5204 | 99.5204 | chlorocebus_sabaeus | ENSCSAP00000002814 | 99.5204 | 99.5204 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 55.1559 | 54.1966 | tupaia_belangeri | ENSTBEP00000010263 | 55.2885 | 54.3269 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 66.6667 | 64.2686 | mesocricetus_auratus | ENSMAUP00000020519 | 98.2332 | 94.6996 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 96.1631 | 92.8058 | phocoena_sinus | ENSPSNP00000008620 | 96.1631 | 92.8058 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 96.4029 | 93.765 | camelus_dromedarius | ENSCDRP00005032606 | 96.4029 | 93.765 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 97.3621 | 95.6835 | carlito_syrichta | ENSTSYP00000027653 | 97.5962 | 95.9135 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 95.4436 | 92.8058 | panthera_tigris_altaica | ENSPTIP00000010365 | 97.0732 | 94.3902 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 97.3621 | 93.765 | rattus_norvegicus | ENSRNOP00000080548 | 97.5962 | 93.9904 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 88.4892 | 84.8921 | prolemur_simus | ENSPSMP00000013415 | 94.6154 | 90.7692 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 96.8825 | 93.5252 | microtus_ochrogaster | ENSMOCP00000003908 | 97.3494 | 93.9759 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 69.5444 | 62.8297 | sorex_araneus | ENSSARP00000005994 | 70.3884 | 63.5922 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 97.8417 | 94.7242 | peromyscus_maniculatus_bairdii | ENSPEMP00000002066 | 98.0769 | 94.9519 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 95.9233 | 92.3261 | bos_mutus | ENSBMUP00000019823 | 95.9233 | 92.3261 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 33.3333 | 31.1751 | mus_spicilegus | ENSMSIP00000016436 | 82.7381 | 77.381 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 97.6019 | 94.2446 | cricetulus_griseus_chok1gshd | ENSCGRP00001021290 | 97.6019 | 94.2446 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 96.8825 | 94.4844 | pteropus_vampyrus | ENSPVAP00000013871 | 96.8825 | 94.4844 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 91.6067 | 83.9329 | vombatus_ursinus | ENSVURP00010016490 | 91.3876 | 83.7321 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 96.1631 | 91.8465 | rhinolophus_ferrumequinum | ENSRFEP00010032128 | 95.933 | 91.6268 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 97.3621 | 95.4436 | neovison_vison | ENSNVIP00000004970 | 97.3621 | 95.4436 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 95.6835 | 92.5659 | bos_taurus | ENSBTAP00000020111 | 95.6835 | 92.5659 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 97.6019 | 94.2446 | heterocephalus_glaber_female | ENSHGLP00000051863 | 73.7319 | 71.1957 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 99.7602 | 99.5204 | rhinopithecus_bieti | ENSRBIP00000029399 | 99.7602 | 99.5204 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 97.3621 | 94.964 | canis_lupus_dingo | ENSCAFP00020006563 | 97.3621 | 94.964 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 96.4029 | 94.2446 | sciurus_vulgaris | ENSSVLP00005022532 | 96.4029 | 94.2446 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 82.2542 | 76.7386 | phascolarctos_cinereus | ENSPCIP00000025664 | 71.9078 | 67.086 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 95.6835 | 92.5659 | procavia_capensis | ENSPCAP00000014777 | 96.1446 | 93.012 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 94.964 | 92.0863 | nannospalax_galili | ENSNGAP00000008233 | 96.3504 | 93.4307 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 95.9233 | 92.5659 | bos_grunniens | ENSBGRP00000011411 | 95.9233 | 92.5659 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 97.3621 | 94.964 | chinchilla_lanigera | ENSCLAP00000018868 | 97.3621 | 94.964 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 88.9688 | 84.6523 | cervus_hanglu_yarkandensis | ENSCHYP00000027149 | 53.8462 | 51.2337 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 94.4844 | 91.6067 | cervus_hanglu_yarkandensis | ENSCHYP00000013696 | 94.4844 | 91.6067 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 96.6427 | 94.2446 | catagonus_wagneri | ENSCWAP00000001000 | 96.6427 | 94.2446 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 96.1631 | 93.5252 | ictidomys_tridecemlineatus | ENSSTOP00000007857 | 95.933 | 93.3014 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 98.801 | 95.4436 | otolemur_garnettii | ENSOGAP00000010958 | 98.801 | 95.4436 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 96.4029 | 94.4844 | cebus_imitator | ENSCCAP00000000260 | 94.8113 | 92.9245 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 95.9233 | 93.0456 | urocitellus_parryii | ENSUPAP00010010118 | 95.6938 | 92.823 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 82.2542 | 78.6571 | moschus_moschiferus | ENSMMSP00000001738 | 81.4727 | 77.9097 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 95.9233 | 92.8058 | moschus_moschiferus | ENSMMSP00000023223 | 95.9233 | 92.8058 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 99.7602 | 99.5204 | rhinopithecus_roxellana | ENSRROP00000042988 | 99.7602 | 99.5204 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 97.6019 | 93.765 | mus_pahari | MGP_PahariEiJ_P0053567 | 97.8365 | 93.9904 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 98.0815 | 95.4436 | propithecus_coquereli | ENSPCOP00000029017 | 98.0815 | 95.4436 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 73.3813 | 59.7122 | caenorhabditis_elegans | Y38A10A.5.1 | 77.4684 | 63.038 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 80.0959 | 68.5851 | ciona_intestinalis | ENSCINP00000016510 | 78.5882 | 67.2941 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 79.1367 | 70.024 | petromyzon_marinus | ENSPMAP00000008682 | 85.0515 | 75.2577 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 62.5899 | 52.518 | callorhinchus_milii | ENSCMIP00000012693 | 62.4402 | 52.3923 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 83.693 | 70.7434 | callorhinchus_milii | ENSCMIP00000025889 | 85.3301 | 72.1271 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 58.9928 | 52.518 | latimeria_chalumnae | ENSLACP00000001920 | 82 | 73 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 87.2902 | 76.259 | lepisosteus_oculatus | ENSLOCP00000011571 | 88.1356 | 76.9976 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 86.3309 | 74.5803 | clupea_harengus | ENSCHAP00000035341 | 84.7059 | 73.1765 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 85.1319 | 73.6211 | danio_rerio | ENSDARP00000092429 | 85.1319 | 73.6211 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 86.0911 | 74.5803 | carassius_auratus | ENSCARP00000053526 | 84.87 | 73.5225 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 84.8921 | 74.8201 | carassius_auratus | ENSCARP00000132752 | 84.0855 | 74.1093 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 84.6523 | 74.5803 | ictalurus_punctatus | ENSIPUP00000002289 | 83.848 | 73.8717 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 62.5899 | 47.482 | esox_lucius | ENSELUP00000024434 | 64.2857 | 48.7685 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 86.3309 | 74.3405 | electrophorus_electricus | ENSEEEP00000013117 | 84.3091 | 72.5995 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 46.5228 | 38.1295 | oncorhynchus_kisutch | ENSOKIP00005068056 | 73.7643 | 60.4563 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 62.1103 | 49.4005 | oncorhynchus_mykiss | ENSOMYP00000118075 | 74.8555 | 59.5376 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 79.6163 | 68.5851 | oncorhynchus_mykiss | ENSOMYP00000008435 | 78.487 | 67.6123 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 84.1727 | 71.223 | oncorhynchus_mykiss | ENSOMYP00000085756 | 82.3944 | 69.7183 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 81.0552 | 68.5851 | oncorhynchus_mykiss | ENSOMYP00000051308 | 82.439 | 69.7561 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 75.2998 | 65.7074 | salmo_salar | ENSSSAP00000079416 | 82.199 | 71.7277 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 80.0959 | 68.8249 | salmo_salar | ENSSSAP00000079447 | 78.2201 | 67.2131 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 76.4988 | 63.3094 | salmo_salar | ENSSSAP00000089025 | 79.5511 | 65.8354 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 79.1367 | 68.3453 | salmo_trutta | ENSSTUP00000103031 | 82.7068 | 71.4286 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 83.693 | 70.2638 | salmo_trutta | ENSSTUP00000106908 | 82.1176 | 68.9412 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 82.9736 | 69.5444 | salmo_trutta | ENSSTUP00000088013 | 81.2207 | 68.0751 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 81.7746 | 70.2638 | salmo_trutta | ENSSTUP00000037541 | 82.5666 | 70.9443 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 85.6115 | 72.6619 | gadus_morhua | ENSGMOP00000015334 | 83.4112 | 70.7944 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 79.3765 | 69.3046 | poecilia_reticulata | ENSPREP00000027157 | 84.6547 | 73.913 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 86.8106 | 75.2998 | xiphophorus_maculatus | ENSXMAP00000003273 | 85.3774 | 74.0566 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 86.3309 | 72.4221 | cyclopterus_lumpus | ENSCLMP00005012877 | 84.3091 | 70.726 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 87.0504 | 75.06 | oreochromis_niloticus | ENSONIP00000002487 | 84.4186 | 72.7907 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 83.9329 | 70.9832 | haplochromis_burtoni | ENSHBUP00000018514 | 80.4598 | 68.046 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 79.3765 | 68.1055 | astatotilapia_calliptera | ENSACLP00000034377 | 84.0102 | 72.0812 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 83.4532 | 72.1823 | sparus_aurata | ENSSAUP00010024705 | 84.058 | 72.7053 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 86.5707 | 74.3405 | lates_calcarifer | ENSLCAP00010057770 | 84.7418 | 72.77 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 22.0623 | 17.506 | sphenodon_punctatus | ENSSPUP00000015365 | 70.7692 | 56.1538 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 76.259 | 69.3046 | laticauda_laticaudata | ENSLLTP00000017009 | 89.3258 | 81.1798 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 85.3717 | 74.3405 | notechis_scutatus | ENSNSUP00000021960 | 84.5606 | 73.6342 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 78.4173 | 71.4628 | pseudonaja_textilis | ENSPTXP00000027450 | 91.8539 | 83.7079 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 86.8106 | 77.9377 | anas_platyrhynchos_platyrhynchos | ENSAPLP00000032112 | 68.1733 | 61.2053 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 89.2086 | 78.1775 | gallus_gallus | ENSGALP00010004657 | 88.7828 | 77.8043 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 75.2998 | 67.6259 | meleagris_gallopavo | ENSMGAP00000002079 | 94.012 | 84.4311 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 24.94 | 21.8225 | serinus_canaria | ENSSCAP00000002133 | 75.9124 | 66.4234 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 77.458 | 70.5036 | parus_major | ENSPMJP00000003678 | 57.8853 | 52.6882 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 21.5827 | 16.307 | anolis_carolinensis | ENSACAP00000009098 | 64.2857 | 48.5714 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 70.2638 | 58.753 | neolamprologus_brichardi | ENSNBRP00000017633 | 84.1954 | 70.4023 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 90.8873 | 81.0552 | podarcis_muralis | ENSPMRP00000036796 | 91.1058 | 81.25 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 43.6451 | 40.048 | geospiza_fortis | ENSGFOP00000004486 | 77.7778 | 71.3675 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 88.2494 | 77.6978 | taeniopygia_guttata | ENSTGUP00000015340 | 89.3204 | 78.6408 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 87.0504 | 74.5803 | erpetoichthys_calabaricus | ENSECRP00000013103 | 86.8421 | 74.4019 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 86.3309 | 73.8609 | kryptolebias_marmoratus | ENSKMAP00000030139 | 82.3799 | 70.4806 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 90.1679 | 81.295 | salvator_merianae | ENSSMRP00000003350 | 90.6024 | 81.6867 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 77.6978 | 69.5444 | oryzias_javanicus | ENSOJAP00000016113 | 85.4881 | 76.5172 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 79.3765 | 68.3453 | amphilophus_citrinellus | ENSACIP00000010879 | 84.0102 | 72.335 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 86.3309 | 72.6619 | dicentrarchus_labrax | ENSDLAP00005082578 | 84.3091 | 70.9602 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 78.6571 | 68.1055 | amphiprion_percula | ENSAPEP00000003876 | 82.4121 | 71.3568 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 91.1271 | 81.295 | gopherus_evgoodei | ENSGEVP00005027181 | 91.3462 | 81.4904 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 87.53 | 78.1775 | chelonoidis_abingdonii | ENSCABP00000003900 | 83.3333 | 74.4292 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 87.0504 | 73.6211 | seriola_dumerili | ENSSDUP00000000032 | 85.4118 | 72.2353 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 85.6115 | 70.5036 | cottoperca_gobio | ENSCGOP00000012701 | 81.5069 | 67.1233 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 87.2902 | 77.6978 | anser_brachyrhynchus | ENSABRP00000011525 | 83.2952 | 74.1419 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 84.6523 | 72.1823 | gasterosteus_aculeatus | ENSGACP00000014621 | 84.4498 | 72.0096 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 84.1727 | 70.5036 | cynoglossus_semilaevis | ENSCSEP00000020843 | 81.4385 | 68.2135 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 86.5707 | 75.2998 | poecilia_formosa | ENSPFOP00000006037 | 84.5433 | 73.5363 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 86.3309 | 73.8609 | sander_lucioperca | ENSSLUP00000024640 | 83.1409 | 71.1316 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 86.3309 | 74.5803 | strigops_habroptila | ENSSHBP00005020242 | 87.5912 | 75.6691 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 85.8513 | 74.1007 | amphiprion_ocellaris | ENSAOCP00000000141 | 83.0626 | 71.6937 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 80.0959 | 73.1415 | terrapene_carolina_triunguis | ENSTMTP00000024864 | 84.3434 | 77.0202 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 78.4173 | 67.6259 | hucho_hucho | ENSHHUP00000071700 | 80.9406 | 69.802 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 82.2542 | 70.5036 | hucho_hucho | ENSHHUP00000075227 | 81.8616 | 70.1671 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 87.0504 | 73.8609 | betta_splendens | ENSBSLP00000017201 | 85.2113 | 72.3005 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 86.5707 | 75.06 | anabas_testudineus | ENSATEP00000009447 | 84.7418 | 73.4742 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 79.6163 | 69.5444 | ciona_savignyi | ENSCSAVP00000001554 | 79.0476 | 69.0476 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 45.0839 | 28.777 | saccharomyces_cerevisiae | YAL058W | 37.4502 | 23.9044 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 86.8106 | 73.1415 | seriola_lalandi_dorsalis | ENSSLDP00000005675 | 85.1765 | 71.7647 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 86.3309 | 73.1415 | labrus_bergylta | ENSLBEP00000008477 | 82.0046 | 69.4761 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 79.3765 | 67.8657 | pundamilia_nyererei | ENSPNYP00000009858 | 83.7975 | 71.6456 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 85.8513 | 73.6211 | stegastes_partitus | ENSSPAP00000024092 | 83.0626 | 71.2297 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 80.5755 | 69.0648 | oncorhynchus_tshawytscha | ENSOTSP00005055753 | 80.5755 | 69.0648 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 76.4988 | 62.8297 | oncorhynchus_tshawytscha | ENSOTSP00005059894 | 79.75 | 65.5 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 86.3309 | 73.3813 | acanthochromis_polyacanthus | ENSAPOP00000032254 | 83.9161 | 71.3287 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 77.2182 | 70.024 | aquila_chrysaetos_chrysaetos | ENSACCP00020018040 | 87.9781 | 79.7814 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 83.693 | 71.4628 | nothobranchius_furzeri | ENSNFUP00015050169 | 77.9018 | 66.5179 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 86.5707 | 74.1007 | cyprinodon_variegatus | ENSCVAP00000019668 | 84.3458 | 72.1963 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 86.0911 | 74.5803 | denticeps_clupeoides | ENSDCDP00000042383 | 82.151 | 71.167 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 38.3693 | 34.5324 | ficedula_albicollis | ENSFALP00000012051 | 84.6561 | 76.1905 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 85.8513 | 74.1007 | hippocampus_comes | ENSHCOP00000001651 | 84.6336 | 73.0496 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 86.0911 | 74.3405 | cyprinus_carpio_carpio | ENSCCRP00000002329 | 85.2732 | 73.6342 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 80.0959 | 72.1823 | cyprinus_carpio_carpio | ENSCCRP00000003280 | 72.9258 | 65.7205 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 81.5348 | 70.7434 | coturnix_japonica | ENSCJPP00005003208 | 88.0829 | 76.4249 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 87.0504 | 75.7794 | poecilia_latipinna | ENSPLAP00000003578 | 85.6132 | 74.5283 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 17.9856 | 16.0671 | sinocyclocheilus_grahami | ENSSGRP00000005666 | 75 | 67 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 26.8585 | 22.0623 | sinocyclocheilus_grahami | ENSSGRP00000006514 | 74.6667 | 61.3333 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 87.2902 | 74.5803 | maylandia_zebra | ENSMZEP00005005697 | 85.0467 | 72.6636 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 75.06 | 67.3861 | tetraodon_nigroviridis | ENSTNIP00000018562 | 84.5946 | 75.9459 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 76.259 | 65.9472 | paramormyrops_kingsleyae | ENSPKIP00000023882 | 85.2547 | 73.7265 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 85.8513 | 74.5803 | scleropages_formosus | ENSSFOP00015024989 | 84.8341 | 73.6967 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 61.3909 | 51.3189 | scleropages_formosus | ENSSFOP00015025833 | 74.8538 | 62.5731 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 80.5755 | 71.7026 | leptobrachium_leishanense | ENSLLEP00000001916 | 87.9581 | 78.2723 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 86.0911 | 73.3813 | scophthalmus_maximus | ENSSMAP00000010589 | 84.4706 | 72 |
| ENSG00000179218 | homo_sapiens | ENSP00000320866 | 55.8753 | 44.3645 | oryzias_melastigma | ENSOMEP00000020614 | 58.25 | 46.25 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0000122 | involved_in negative regulation of transcription by RNA polymerase II | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0001669 | located_in acrosomal vesicle | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0001849 | enables complement component C1q complex binding | 2 | IPI,TAS | Homo_sapiens(9606) | Function |
| GO:0002502 | involved_in peptide antigen assembly with MHC class I protein complex | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0003677 | enables DNA binding | 1 | NAS | Homo_sapiens(9606) | Function |
| GO:0003723 | enables RNA binding | 1 | HDA | Homo_sapiens(9606) | Function |
| GO:0003729 | enables mRNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0005049 | enables nuclear export signal receptor activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0005178 | enables integrin binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005506 | enables iron ion binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0005509 | enables calcium ion binding | 5 | IBA,IDA,IMP,ISS,TAS | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005576 | located_in extracellular region | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0005615 | located_in extracellular space | 2 | HDA,IMP | Homo_sapiens(9606) | Component |
| GO:0005634 | located_in nucleus | 2 | HDA,IDA | Homo_sapiens(9606) | Component |
| GO:0005635 | located_in nuclear envelope | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005737 | located_in cytoplasm | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005783 | located_in endoplasmic reticulum | 2 | IDA,TAS | Homo_sapiens(9606) | Component |
| GO:0005788 | located_in endoplasmic reticulum lumen | 2 | IDA,TAS | Homo_sapiens(9606) | Component |
| GO:0005789 | is_active_in endoplasmic reticulum membrane | 1 | IBA | Homo_sapiens(9606) | Component |
| GO:0005790 | located_in smooth endoplasmic reticulum | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0005829 | located_in cytosol | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005844 | part_of polysome | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005925 | located_in focal adhesion | 1 | HDA | Homo_sapiens(9606) | Component |
| GO:0006355 | involved_in regulation of DNA-templated transcription | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0006457 | involved_in protein folding | 2 | IBA,TAS | Homo_sapiens(9606) | Process |
| GO:0006611 | involved_in protein export from nucleus | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0006874 | involved_in intracellular calcium ion homeostasis | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0007283 | involved_in spermatogenesis | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0008270 | enables zinc ion binding | 1 | TAS | Homo_sapiens(9606) | Function |
| GO:0008284 | involved_in positive regulation of cell population proliferation | 1 | IGI | Homo_sapiens(9606) | Process |
| GO:0009410 | involved_in response to xenobiotic stimulus | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0009897 | located_in external side of plasma membrane | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0009986 | located_in cell surface | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0010595 | involved_in positive regulation of endothelial cell migration | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0010628 | involved_in positive regulation of gene expression | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0016020 | located_in membrane | 2 | HDA,IDA | Homo_sapiens(9606) | Component |
| GO:0017148 | involved_in negative regulation of translation | 2 | IDA,TAS | Homo_sapiens(9606) | Process |
| GO:0022417 | involved_in protein maturation by protein folding | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0030246 | enables carbohydrate binding | 1 | TAS | Homo_sapiens(9606) | Function |
| GO:0030433 | involved_in ubiquitin-dependent ERAD pathway | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0030670 | located_in phagocytic vesicle membrane | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0030866 | involved_in cortical actin cytoskeleton organization | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0031625 | enables ubiquitin protein ligase binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0032355 | involved_in response to estradiol | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0033018 | located_in sarcoplasmic reticulum lumen | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0033116 | located_in endoplasmic reticulum-Golgi intermediate compartment membrane | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0033144 | involved_in negative regulation of intracellular steroid hormone receptor signaling pathway | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0033574 | involved_in response to testosterone | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0034504 | involved_in protein localization to nucleus | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0034975 | involved_in protein folding in endoplasmic reticulum | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0040020 | involved_in regulation of meiotic nuclear division | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0042277 | enables peptide binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0042562 | enables hormone binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0042824 | part_of MHC class I peptide loading complex | 2 | IDA,ISS | Homo_sapiens(9606) | Component |
| GO:0042921 | involved_in glucocorticoid receptor signaling pathway | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0042981 | involved_in regulation of apoptotic process | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0044183 | enables protein folding chaperone | 1 | TAS | Homo_sapiens(9606) | Function |
| GO:0044194 | located_in cytolytic granule | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0044322 | located_in endoplasmic reticulum quality control compartment | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0045665 | involved_in negative regulation of neuron differentiation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0045787 | acts_upstream_of positive regulation of cell cycle | 1 | IGI | Homo_sapiens(9606) | Process |
| GO:0045892 | involved_in negative regulation of DNA-templated transcription | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0048387 | involved_in negative regulation of retinoic acid receptor signaling pathway | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0048471 | located_in perinuclear region of cytoplasm | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0050681 | enables nuclear androgen receptor binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0050766 | involved_in positive regulation of phagocytosis | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0050821 | involved_in protein stabilization | 2 | ISS,TAS | Homo_sapiens(9606) | Process |
| GO:0051082 | enables unfolded protein binding | 1 | TAS | Homo_sapiens(9606) | Function |
| GO:0051087 | enables protein-folding chaperone binding | 1 | TAS | Homo_sapiens(9606) | Function |
| GO:0051208 | involved_in sequestering of calcium ion | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0055007 | involved_in cardiac muscle cell differentiation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0060473 | located_in cortical granule | 1 | ISS | Homo_sapiens(9606) | Component |
| GO:0061912 | involved_in selective autophagy | 1 | EXP | Homo_sapiens(9606) | Process |
| GO:0062023 | located_in collagen-containing extracellular matrix | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0070062 | located_in extracellular exosome | 1 | HDA | Homo_sapiens(9606) | Component |
| GO:0071257 | involved_in cellular response to electrical stimulus | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0071285 | involved_in cellular response to lithium ion | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0071682 | located_in endocytic vesicle lumen | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0090398 | involved_in cellular senescence | 1 | IGI | Homo_sapiens(9606) | Process |
| GO:0098553 | located_in lumenal side of endoplasmic reticulum membrane | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0098586 | involved_in cellular response to virus | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:1900026 | involved_in positive regulation of substrate adhesion-dependent cell spreading | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:1901164 | involved_in negative regulation of trophoblast cell migration | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:1901224 | involved_in positive regulation of NIK/NF-kappaB signaling | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:1903416 | involved_in response to glycoside | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:2000510 | involved_in positive regulation of dendritic cell chemotaxis | 1 | IMP | Homo_sapiens(9606) | Process |