RBP Type
Non-canonical_RBPs
Diseases
Drug
N.A.
Main interacting RNAs
N.A.
Moonlighting functions
Metabolic enzyme
Localizations
N.A.
BulkPerturb-seq
Ensembl ID ENSG00000169919 Gene ID
2990 Accession
4696
Symbol
GUSB
Alias
BG;MPS7
Full Name
glucuronidase beta
Status
Confidence
Length
21532 bases
Strand
Minus strand
Position
7 : 65960684 - 65982215
RNA binding domain
N.A.
Summary
This gene encodes a hydrolase that degrades glycosaminoglycans, including heparan sulfate, dermatan sulfate, and chondroitin-4,6-sulfate. The enzyme forms a homotetramer that is localized to the lysosome. Mutations in this gene result in mucopolysaccharidosis type VII. Alternative splicing results in multiple transcript variants. There are many pseudogenes of this locus in the human genome.[provided by RefSeq, May 2014]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
Name
Transcript ID
bp
Protein
Translation ID
GUSB-201
ENST00000304895
2199
651aa
ENSP00000302728
GUSB-209
ENST00000466883
2555
No protein
-
GUSB-205
ENST00000447929
1983
144aa
ENSP00000411262
GUSB-203
ENST00000430730
1859
134aa
ENSP00000411859
GUSB-202
ENST00000421103
1742
505aa
ENSP00000391390
GUSB-206
ENST00000461622
491
No protein
-
GUSB-207
ENST00000462371
820
No protein
-
GUSB-208
ENST00000465785
625
No protein
-
GUSB-213
ENST00000479038
452
No protein
-
GUSB-214
ENST00000489482
519
No protein
-
GUSB-210
ENST00000475316
505
No protein
-
GUSB-211
ENST00000476486
776
No protein
-
GUSB-204
ENST00000446111
933
144aa
ENSP00000416793
GUSB-212
ENST00000478118
571
No protein
-
Pathway ID
Pathway Name
Source
ensgID
Trait
pValue
Pubmed ID
27800 Blood Glucose 5.3950000E-005 - 34988 Platelet Function Tests 2.1311100E-005 -
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
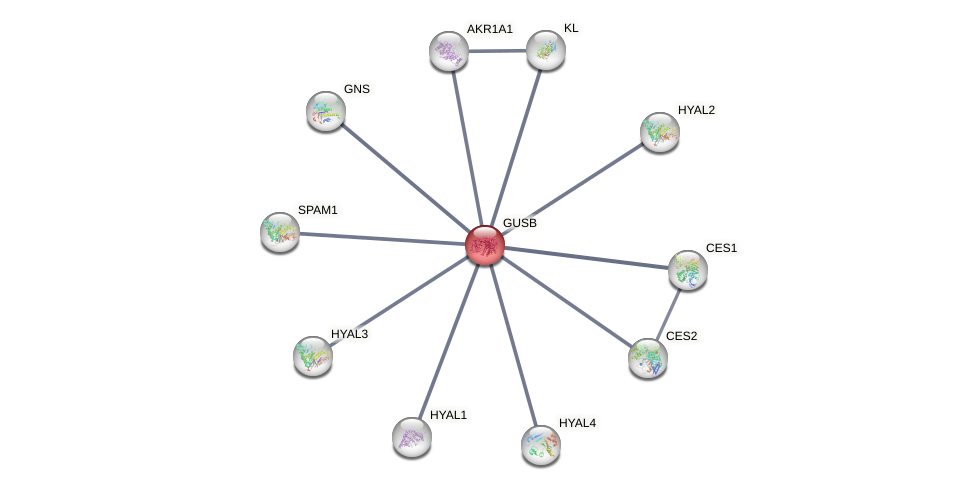
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0004566 enables beta-glucuronidase activity 2 IBA,TAS Homo_sapiens(9606) Function GO:0005102 enables signaling receptor binding 2 IBA,IPI Homo_sapiens(9606) Function GO:0005576 located_in extracellular region 1 TAS Homo_sapiens(9606) Component GO:0005615 is_active_in extracellular space 2 IBA,IDA Homo_sapiens(9606) Component GO:0005975 involved_in carbohydrate metabolic process 1 IEA Homo_sapiens(9606) Process GO:0006027 involved_in glycosaminoglycan catabolic process 1 TAS Homo_sapiens(9606) Process GO:0016020 located_in membrane 1 HDA Homo_sapiens(9606) Component GO:0019391 involved_in glucuronoside catabolic process 1 IBA Homo_sapiens(9606) Process GO:0019904 enables protein domain specific binding 1 IPI Homo_sapiens(9606) Function GO:0030200 involved_in heparan sulfate proteoglycan catabolic process 1 IEA Homo_sapiens(9606) Process GO:0030207 involved_in chondroitin sulfate catabolic process 1 IEA Homo_sapiens(9606) Process GO:0030214 involved_in hyaluronan catabolic process 1 IEA Homo_sapiens(9606) Process GO:0030246 enables carbohydrate binding 1 IBA Homo_sapiens(9606) Function GO:0035578 located_in azurophil granule lumen 1 TAS Homo_sapiens(9606) Component GO:0043202 located_in lysosomal lumen 1 TAS Homo_sapiens(9606) Component GO:0043231 located_in intracellular membrane-bounded organelle 1 IDA Homo_sapiens(9606) Component GO:0070062 located_in extracellular exosome 1 HDA Homo_sapiens(9606) Component GO:1904813 located_in ficolin-1-rich granule lumen 1 TAS Homo_sapiens(9606) Component
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.