RBP Type
Non-canonical_RBPs
Diseases
Flaviviruses SARS-COV-2
Drug
N.A.
Main interacting RNAs
N.A.
Moonlighting functions
E3 ligase
Localizations
Stress granucle
BulkPerturb-seq
Ensembl ID ENSG00000169871 Gene ID
81844 Accession
19028
Symbol
TRIM56
Alias
RNF109
Full Name
tripartite motif containing 56
Status
Confidence
Length
12487 bases
Strand
Plus strand
Position
7 : 101085481 - 101097967
RNA binding domain
N.A.
Summary
Enables RNA binding activity. Predicted to be involved in several processes, including defense response to other organism; positive regulation of macromolecule metabolic process; and protein K63-linked ubiquitination. Predicted to be located in cytoplasm. Predicted to be active in chromatin and nucleoplasm. [provided by Alliance of Genome Resources, Apr 2022]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
31251739 The E3 ligase TRIM56 is a host restriction factor of Zika virus and depends on its RNA-binding activity but not miRNA regulation, for antiviral function.
Darong Yang
2019-06-01
PLoS neglected tropical diseases
36902478 The Functions of TRIM56 in Antiviral Innate Immunity and Tumorigenesis.
Lin Fu
2023-03-06
International journal of molecular sciences
32738036 Molecular and biological functions of TRIM-NHL RNA-binding proteins.
P Robert Connacher
2021-03-01
Wiley interdisciplinary reviews. RNA
34899698 Roles of Emerging RNA-Binding Activity of cGAS in Innate Antiviral Response.
Yuying Ma
2021-01-01
Frontiers in immunology
32829025 The antiviral activities of TRIM proteins.
Lennart Koepke
2021-02-01
Current opinion in microbiology
29215570 Host Cell Restriction Factors that Limit Influenza A Infection.
Fernando Villalón-Letelier
2017-12-07
Viruses
36381143 Comparative structural analyses of the NHL domains from the human E3 ligase TRIM-NHL family.
Apirat Chaikuad
2022-11-01
IUCrJ
35062293 Impaired Antiviral Responses to Extracellular Double-Stranded RNA and Cytosolic DNA, but Not to Interferon-α Stimulation, in TRIM56-Deficient Cells.
Dang Wang
2022-01-05
Viruses
Name
Transcript ID
bp
Protein
Translation ID
TRIM56-201
ENST00000306085
10910
755aa
ENSP00000305161
TRIM56-202
ENST00000412507
1129
305aa
ENSP00000404186
TRIM56-203
ENST00000467847
581
65aa
ENSP00000486217
TRIM56-204
ENST00000487252
735
No protein
-
Pathway ID
Pathway Name
Source
ensgID
Trait
pValue
Pubmed ID
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
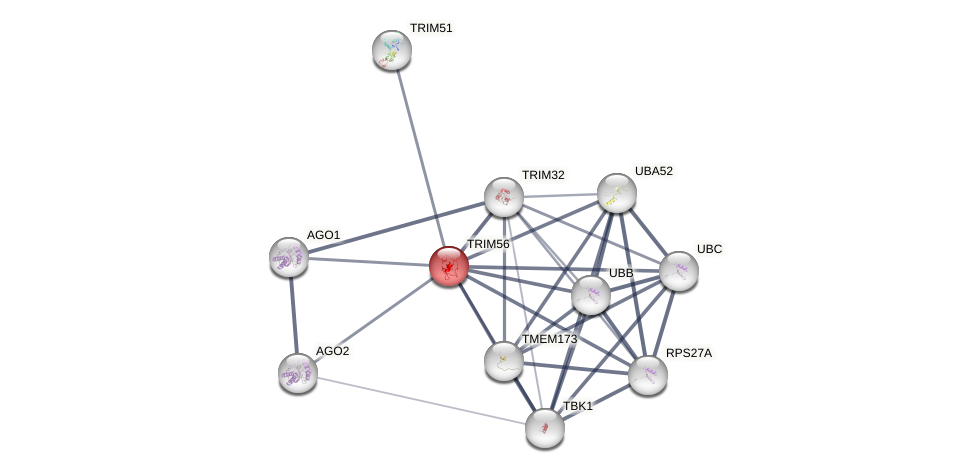
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
ENSG00000169871 homo_sapiens ENSP00000366404 25.1462 16.5692 homo_sapiens ENSP00000305161 17.0861 11.2583 ENSG00000169871 homo_sapiens ENSP00000256649 19.8276 11.8966 homo_sapiens ENSP00000305161 15.2318 9.13907 ENSG00000169871 homo_sapiens ENSP00000254816 19.4357 13.4796 homo_sapiens ENSP00000305161 16.4238 11.3907 ENSG00000169871 homo_sapiens ENSP00000496301 19.7822 11.7967 homo_sapiens ENSP00000305161 14.4371 8.60927 ENSG00000169871 homo_sapiens ENSP00000518164 24.2038 12.9512 homo_sapiens ENSP00000305161 15.0993 8.07947 ENSG00000169871 homo_sapiens ENSP00000421142 24.8408 13.3758 homo_sapiens ENSP00000305161 15.4967 8.34437 ENSG00000169871 homo_sapiens ENSP00000423934 15.6425 9.63687 homo_sapiens ENSP00000305161 14.8344 9.13907 ENSG00000169871 homo_sapiens ENSP00000365884 28.172 15.6989 homo_sapiens ENSP00000305161 17.351 9.66887 ENSG00000169871 homo_sapiens ENSP00000351250 12.4224 6.29991 homo_sapiens ENSP00000305161 18.543 9.40397 ENSG00000169871 homo_sapiens ENSP00000262294 12.9668 7.57261 homo_sapiens ENSP00000305161 16.5563 9.66887 ENSG00000169871 homo_sapiens ENSP00000327994 21.1499 13.7577 homo_sapiens ENSP00000305161 13.6424 8.87417 ENSG00000169871 homo_sapiens ENSP00000254436 28.8421 16.8421 homo_sapiens ENSP00000305161 18.1457 10.596 ENSG00000169871 homo_sapiens ENSP00000343129 17.3469 9.69388 homo_sapiens ENSP00000305161 13.5099 7.54967 ENSG00000169871 homo_sapiens ENSP00000507131 11.6981 7.04402 homo_sapiens ENSP00000305161 12.3179 7.41722 ENSG00000169871 homo_sapiens ENSP00000355613 10.9834 6.3857 homo_sapiens ENSP00000305161 11.3907 6.62252 ENSG00000169871 homo_sapiens ENSP00000474678 21.0177 11.7257 homo_sapiens ENSP00000305161 12.5828 7.01987 ENSG00000169871 homo_sapiens ENSP00000485097 21.0177 11.7257 homo_sapiens ENSP00000305161 12.5828 7.01987 ENSG00000169871 homo_sapiens ENSP00000402414 20.9821 14.2857 homo_sapiens ENSP00000305161 6.22517 4.23841 ENSG00000169871 homo_sapiens ENSP00000291416 28 16.6316 homo_sapiens ENSP00000305161 17.6159 10.4636 ENSG00000169871 homo_sapiens ENSP00000369373 25.9635 15.6187 homo_sapiens ENSP00000305161 16.9536 10.1987 ENSG00000169871 homo_sapiens ENSP00000268058 16.0998 8.61678 homo_sapiens ENSP00000305161 18.8079 10.0662 ENSG00000169871 homo_sapiens ENSP00000318615 23.6 15.2 homo_sapiens ENSP00000305161 7.81457 5.03311 ENSG00000169871 homo_sapiens ENSP00000285805 23.6 15.2 homo_sapiens ENSP00000305161 7.81457 5.03311 ENSG00000169871 homo_sapiens ENSP00000474003 22.6667 12 homo_sapiens ENSP00000305161 13.5099 7.15232 ENSG00000169871 homo_sapiens ENSP00000312675 23.8994 13.4172 homo_sapiens ENSP00000305161 15.0993 8.47682 ENSG00000169871 homo_sapiens ENSP00000299413 14.8256 7.55814 homo_sapiens ENSP00000305161 6.75497 3.44371 ENSG00000169871 homo_sapiens ENSP00000349596 26.2366 14.8387 homo_sapiens ENSP00000305161 16.1589 9.13907 ENSG00000169871 homo_sapiens ENSP00000284551 25 14.9573 homo_sapiens ENSP00000305161 15.4967 9.27152 ENSG00000169871 homo_sapiens ENSP00000367424 25.7985 12.7764 homo_sapiens ENSP00000305161 13.9073 6.88742 ENSG00000169871 homo_sapiens ENSP00000402595 22.541 12.7049 homo_sapiens ENSP00000305161 14.5695 8.21192 ENSG00000169871 homo_sapiens ENSP00000495413 7.75862 4.23851 homo_sapiens ENSP00000305161 14.3046 7.81457 ENSG00000169871 homo_sapiens ENSP00000340797 18.414 10.4839 homo_sapiens ENSP00000305161 18.1457 10.3311 ENSG00000169871 homo_sapiens ENSP00000323889 19.0476 10.1587 homo_sapiens ENSP00000305161 15.894 8.47682 ENSG00000169871 homo_sapiens ENSP00000492719 23.3184 13.6771 homo_sapiens ENSP00000305161 13.7748 8.07947 ENSG00000169871 homo_sapiens ENSP00000272395 23.3184 13.6771 homo_sapiens ENSP00000305161 13.7748 8.07947 ENSG00000169871 homo_sapiens ENSP00000340507 13.619 7.33333 homo_sapiens ENSP00000305161 18.9404 10.1987 ENSG00000169871 homo_sapiens ENSP00000379800 27.8689 17.0082 homo_sapiens ENSP00000305161 18.0132 10.9934 ENSG00000169871 homo_sapiens ENSP00000481815 24 12.6667 homo_sapiens ENSP00000305161 14.3046 7.54967 ENSG00000169871 homo_sapiens ENSP00000332969 24.7216 13.5857 homo_sapiens ENSP00000305161 14.702 8.07947 ENSG00000169871 homo_sapiens ENSP00000483764 24.4989 13.8085 homo_sapiens ENSP00000305161 14.5695 8.21192 ENSG00000169871 homo_sapiens ENSP00000332284 26.8 15.2 homo_sapiens ENSP00000305161 17.7483 10.0662 ENSG00000169871 homo_sapiens ENSP00000244360 26.9886 17.3295 homo_sapiens ENSP00000305161 12.5828 8.07947 ENSG00000169871 homo_sapiens ENSP00000491320 26.2821 13.0342 homo_sapiens ENSP00000305161 16.2914 8.07947 ENSG00000169871 homo_sapiens ENSP00000323913 18.4307 10.7664 homo_sapiens ENSP00000305161 13.3775 7.81457 ENSG00000169871 homo_sapiens ENSP00000369415 23.4637 14.8045 homo_sapiens ENSP00000305161 11.1258 7.01987 ENSG00000169871 homo_sapiens ENSP00000363298 25.8706 14.4279 homo_sapiens ENSP00000305161 13.7748 7.68212 ENSG00000169871 homo_sapiens ENSP00000379826 29.0698 15.5039 homo_sapiens ENSP00000305161 9.93377 5.29801 ENSG00000169871 homo_sapiens ENSP00000320869 19.8413 11.746 homo_sapiens ENSP00000305161 16.5563 9.80132 ENSG00000169871 homo_sapiens ENSP00000508553 18.4669 11.4983 homo_sapiens ENSP00000305161 7.01987 4.37086 ENSG00000169871 homo_sapiens ENSP00000408292 20.9801 12.098 homo_sapiens ENSP00000305161 18.1457 10.4636 ENSG00000169871 homo_sapiens ENSP00000262843 14.5578 8.84354 homo_sapiens ENSP00000305161 14.1722 8.60927 ENSG00000169871 homo_sapiens ENSP00000344208 42.3077 26.4706 homo_sapiens ENSP00000305161 24.7682 15.4967 ENSG00000169871 homo_sapiens ENSP00000373272 15.3226 8.64055 homo_sapiens ENSP00000305161 17.6159 9.93377 ENSG00000169871 homo_sapiens ENSP00000397073 28.8981 17.0478 homo_sapiens ENSP00000305161 18.4106 10.8609 ENSG00000169871 homo_sapiens ENSP00000497185 23.7589 12.9433 homo_sapiens ENSP00000305161 17.7483 9.66887 ENSG00000169871 homo_sapiens ENSP00000286349 16.8741 9.68188 homo_sapiens ENSP00000305161 16.1589 9.27152 ENSG00000169871 homo_sapiens ENSP00000339659 18.0285 10.1167 homo_sapiens ENSP00000305161 18.4106 10.3311 ENSG00000169871 homo_sapiens ENSP00000355659 24.9476 15.9329 homo_sapiens ENSP00000305161 15.7616 10.0662 ENSG00000169871 homo_sapiens ENSP00000253024 14.012 7.78443 homo_sapiens ENSP00000305161 15.4967 8.60927 ENSG00000169871 homo_sapiens ENSP00000274773 22.1135 13.8943 homo_sapiens ENSP00000305161 14.9669 9.40397 ENSG00000169871 homo_sapiens ENSP00000507413 25.6293 12.357 homo_sapiens ENSP00000305161 14.8344 7.15232 ENSG00000169871 homo_sapiens ENSP00000410446 20.9648 11.8738 homo_sapiens ENSP00000305161 14.9669 8.47682 ENSG00000169871 homo_sapiens ENSP00000301924 26.572 15.213 homo_sapiens ENSP00000305161 17.351 9.93377 ENSG00000169871 homo_sapiens ENSP00000363390 24.0793 13.881 homo_sapiens ENSP00000305161 11.2583 6.49007 ENSG00000169871 homo_sapiens ENSP00000369299 24.2972 13.8554 homo_sapiens ENSP00000305161 16.0265 9.13907 ENSG00000169871 homo_sapiens ENSP00000312678 15.8921 9.29535 homo_sapiens ENSP00000305161 14.0397 8.21192 ENSG00000169871 homo_sapiens ENSP00000327738 27.7778 16.8803 homo_sapiens ENSP00000305161 17.2185 10.4636 ENSG00000169871 homo_sapiens ENSP00000330216 20.354 11.7257 homo_sapiens ENSP00000305161 12.1854 7.01987 ENSG00000169871 homo_sapiens ENSP00000388299 20.7965 11.2832 homo_sapiens ENSP00000305161 12.4503 6.75497 ENSG00000169871 homo_sapiens ENSP00000327604 20.7965 11.2832 homo_sapiens ENSP00000305161 12.4503 6.75497 ENSG00000169871 homo_sapiens ENSP00000269383 19.5358 12.766 homo_sapiens ENSP00000305161 13.3775 8.74172 ENSG00000169871 homo_sapiens ENSP00000300747 26.8041 15.6701 homo_sapiens ENSP00000305161 17.2185 10.0662 ENSG00000169871 homo_sapiens ENSP00000395086 22.3451 13.2743 homo_sapiens ENSP00000305161 13.3775 7.94702 ENSG00000169871 homo_sapiens ENSP00000497050 21.0177 12.1681 homo_sapiens ENSP00000305161 12.5828 7.28477 ENSG00000169871 homo_sapiens ENSP00000275736 22.5738 11.8143 homo_sapiens ENSP00000305161 14.1722 7.41722 ENSG00000169871 homo_sapiens ENSP00000334657 16.7325 9.74967 homo_sapiens ENSP00000305161 16.8212 9.80132 ENSG00000169871 homo_sapiens ENSP00000355437 28.3951 17.0782 homo_sapiens ENSP00000305161 18.2781 10.9934 ENSG00000169871 homo_sapiens ENSP00000365924 24 12.2353 homo_sapiens ENSP00000305161 13.5099 6.88742 ENSG00000169871 homo_sapiens ENSP00000219596 11.0115 5.12164 homo_sapiens ENSP00000305161 11.3907 5.29801 ENSG00000169871 homo_sapiens ENSP00000369440 23.4496 12.7907 homo_sapiens ENSP00000305161 16.0265 8.74172
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
ENSG00000169871 homo_sapiens ENSP00000305161 98.6755 98.1457 nomascus_leucogenys ENSNLEP00000043213 98.6755 98.1457 ENSG00000169871 homo_sapiens ENSP00000305161 99.6026 99.3377 pan_paniscus ENSPPAP00000001701 99.6026 99.3377 ENSG00000169871 homo_sapiens ENSP00000305161 99.2053 98.543 pongo_abelii ENSPPYP00000019548 99.2053 98.543 ENSG00000169871 homo_sapiens ENSP00000305161 99.6026 99.3377 pan_troglodytes ENSPTRP00000033419 99.6026 99.3377 ENSG00000169871 homo_sapiens ENSP00000305161 99.4702 98.9404 gorilla_gorilla ENSGGOP00000000399 99.4702 98.9404 ENSG00000169871 homo_sapiens ENSP00000305161 86.8874 80.5298 dasypus_novemcinctus ENSDNOP00000029719 85.6397 79.3734 ENSG00000169871 homo_sapiens ENSP00000305161 31.2583 28.6093 erinaceus_europaeus ENSEEUP00000012875 87.0849 79.7048 ENSG00000169871 homo_sapiens ENSP00000305161 26.3576 25.5629 echinops_telfairi ENSETEP00000002470 91.7051 88.9401 ENSG00000169871 homo_sapiens ENSP00000305161 96.5563 94.5695 callithrix_jacchus ENSCJAP00000029370 96.5563 94.5695 ENSG00000169871 homo_sapiens ENSP00000305161 98.4106 97.2185 cercocebus_atys ENSCATP00000003192 88.2423 87.1734 ENSG00000169871 homo_sapiens ENSP00000305161 33.5099 33.245 macaca_fascicularis ENSMFAP00000014634 99.2157 98.4314 ENSG00000169871 homo_sapiens ENSP00000305161 98.4106 97.2185 macaca_mulatta ENSMMUP00000022856 98.4106 97.2185 ENSG00000169871 homo_sapiens ENSP00000305161 98.4106 97.2185 macaca_nemestrina ENSMNEP00000029482 98.4106 97.2185 ENSG00000169871 homo_sapiens ENSP00000305161 98.4106 97.0861 papio_anubis ENSPANP00000060734 98.4106 97.0861 ENSG00000169871 homo_sapiens ENSP00000305161 97.6159 96.2914 mandrillus_leucophaeus ENSMLEP00000018457 98.2667 96.9333 ENSG00000169871 homo_sapiens ENSP00000305161 92.4503 87.947 tursiops_truncatus ENSTTRP00000000716 92.4503 87.947 ENSG00000169871 homo_sapiens ENSP00000305161 94.5695 90.4636 vulpes_vulpes ENSVVUP00000032563 73.3813 70.1953 ENSG00000169871 homo_sapiens ENSP00000305161 86.8874 80 mus_spretus MGP_SPRETEiJ_P0074265 89.3733 82.2888 ENSG00000169871 homo_sapiens ENSP00000305161 95.2318 90.4636 equus_asinus ENSEASP00005037878 93.2555 88.5863 ENSG00000169871 homo_sapiens ENSP00000305161 90.7285 86.4901 capra_hircus ENSCHIP00000015714 93.4516 89.0859 ENSG00000169871 homo_sapiens ENSP00000305161 89.8013 84.7682 loxodonta_africana ENSLAFP00000026313 92.1196 86.9565 ENSG00000169871 homo_sapiens ENSP00000305161 94.3046 89.9338 panthera_leo ENSPLOP00000019584 92.4675 88.1818 ENSG00000169871 homo_sapiens ENSP00000305161 92.053 87.4172 bos_indicus_hybrid ENSBIXP00005006061 92.053 87.4172 ENSG00000169871 homo_sapiens ENSP00000305161 84.3709 79.4702 oryctolagus_cuniculus ENSOCUP00000041178 91 85.7143 ENSG00000169871 homo_sapiens ENSP00000305161 95.0993 90.1987 equus_caballus ENSECAP00000073060 93.1258 88.3269 ENSG00000169871 homo_sapiens ENSP00000305161 94.9669 90.7285 canis_lupus_familiaris ENSCAFP00845000776 92.9961 88.8457 ENSG00000169871 homo_sapiens ENSP00000305161 33.3775 32.053 marmota_marmota_marmota ENSMMMP00000013136 98.8235 94.902 ENSG00000169871 homo_sapiens ENSP00000305161 92.5828 87.8146 balaenoptera_musculus ENSBMSP00010014936 92.5828 87.8146 ENSG00000169871 homo_sapiens ENSP00000305161 91.1258 87.2848 cavia_porcellus ENSCPOP00000023114 93.9891 90.0273 ENSG00000169871 homo_sapiens ENSP00000305161 94.3046 89.8013 felis_catus ENSFCAP00000055879 92.4675 88.0519 ENSG00000169871 homo_sapiens ENSP00000305161 94.702 90.9934 ursus_americanus ENSUAMP00000010414 92.7367 89.1051 ENSG00000169871 homo_sapiens ENSP00000305161 61.5894 59.0728 bison_bison_bison ENSBBBP00000022726 61.5894 59.0728 ENSG00000169871 homo_sapiens ENSP00000305161 92.4503 87.8146 monodon_monoceros ENSMMNP00015011893 92.4503 87.8146 ENSG00000169871 homo_sapiens ENSP00000305161 92.7152 88.2119 sus_scrofa ENSSSCP00000054250 92.7152 88.2119 ENSG00000169871 homo_sapiens ENSP00000305161 91.1258 87.8146 microcebus_murinus ENSMICP00000027826 92.2252 88.874 ENSG00000169871 homo_sapiens ENSP00000305161 90.8609 86.8874 octodon_degus ENSODEP00000022192 93.588 89.4952 ENSG00000169871 homo_sapiens ENSP00000305161 86.8874 81.1921 jaculus_jaculus ENSJJAP00000006070 90.2338 84.3191 ENSG00000169871 homo_sapiens ENSP00000305161 92.1854 87.1523 ovis_aries_rambouillet ENSOARP00020018544 82.9559 78.4267 ENSG00000169871 homo_sapiens ENSP00000305161 89.8013 86.2252 ursus_maritimus ENSUMAP00000015466 94.6927 90.9218 ENSG00000169871 homo_sapiens ENSP00000305161 92.3179 87.6821 delphinapterus_leucas ENSDLEP00000011245 92.3179 87.6821 ENSG00000169871 homo_sapiens ENSP00000305161 92.3179 87.947 physeter_catodon ENSPCTP00005014652 92.3179 87.947 ENSG00000169871 homo_sapiens ENSP00000305161 87.0199 80.3974 mus_caroli MGP_CAROLIEiJ_P0071454 89.5095 82.6975 ENSG00000169871 homo_sapiens ENSP00000305161 87.1523 80.3974 mus_musculus ENSMUSP00000058109 89.6458 82.6975 ENSG00000169871 homo_sapiens ENSP00000305161 84.3709 76.9536 dipodomys_ordii ENSDORP00000019449 86.5489 78.9402 ENSG00000169871 homo_sapiens ENSP00000305161 94.3046 90.4636 mustela_putorius_furo ENSMPUP00000019638 92.3476 88.5863 ENSG00000169871 homo_sapiens ENSP00000305161 95.3642 92.8477 saimiri_boliviensis_boliviensis ENSSBOP00000011283 96.2567 93.7166 ENSG00000169871 homo_sapiens ENSP00000305161 98.4106 97.0861 chlorocebus_sabaeus ENSCSAP00000018990 98.4106 97.0861 ENSG00000169871 homo_sapiens ENSP00000305161 86.755 79.6026 mesocricetus_auratus ENSMAUP00000012528 88.9946 81.6576 ENSG00000169871 homo_sapiens ENSP00000305161 92.1854 87.5497 phocoena_sinus ENSPSNP00000010188 92.1854 87.5497 ENSG00000169871 homo_sapiens ENSP00000305161 92.7152 87.4172 camelus_dromedarius ENSCDRP00005021442 93.7082 88.3534 ENSG00000169871 homo_sapiens ENSP00000305161 33.1126 32.1854 carlito_syrichta ENSTSYP00000015193 98.0392 95.2941 ENSG00000169871 homo_sapiens ENSP00000305161 87.4172 81.0596 rattus_norvegicus ENSRNOP00000095109 89.9183 83.3787 ENSG00000169871 homo_sapiens ENSP00000305161 93.7748 91.1258 prolemur_simus ENSPSMP00000014465 93.7748 91.1258 ENSG00000169871 homo_sapiens ENSP00000305161 87.2848 81.0596 microtus_ochrogaster ENSMOCP00000003977 89.782 83.3787 ENSG00000169871 homo_sapiens ENSP00000305161 87.1523 80.1325 peromyscus_maniculatus_bairdii ENSPEMP00000031995 88.9189 81.7568 ENSG00000169871 homo_sapiens ENSP00000305161 92.053 87.4172 bos_mutus ENSBMUP00000007799 92.053 87.4172 ENSG00000169871 homo_sapiens ENSP00000305161 87.1523 80.3974 mus_spicilegus ENSMSIP00000011717 89.6458 82.6975 ENSG00000169871 homo_sapiens ENSP00000305161 89.0066 86.3576 pteropus_vampyrus ENSPVAP00000009030 89.0066 86.3576 ENSG00000169871 homo_sapiens ENSP00000305161 92.3179 88.7417 myotis_lucifugus ENSMLUP00000009011 92.3179 88.7417 ENSG00000169871 homo_sapiens ENSP00000305161 94.5695 90.3311 rhinolophus_ferrumequinum ENSRFEP00010009022 93.578 89.384 ENSG00000169871 homo_sapiens ENSP00000305161 94.3046 90.1987 neovison_vison ENSNVIP00000002127 92.3476 88.3269 ENSG00000169871 homo_sapiens ENSP00000305161 98.0132 96.5563 rhinopithecus_bieti ENSRBIP00000005024 98.0132 96.5563 ENSG00000169871 homo_sapiens ENSP00000305161 94.9669 90.7285 canis_lupus_dingo ENSCAFP00020016614 92.9961 88.8457 ENSG00000169871 homo_sapiens ENSP00000305161 92.8477 89.1391 sciurus_vulgaris ENSSVLP00005022140 92.7249 89.0212 ENSG00000169871 homo_sapiens ENSP00000305161 76.8212 71.1258 nannospalax_galili ENSNGAP00000011390 89.2308 82.6154 ENSG00000169871 homo_sapiens ENSP00000305161 92.053 87.4172 bos_grunniens ENSBGRP00000040555 92.053 87.4172 ENSG00000169871 homo_sapiens ENSP00000305161 90.9934 88.2119 chinchilla_lanigera ENSCLAP00000022188 93.8525 90.9836 ENSG00000169871 homo_sapiens ENSP00000305161 92.3179 87.6821 cervus_hanglu_yarkandensis ENSCHYP00000022151 92.3179 87.6821 ENSG00000169871 homo_sapiens ENSP00000305161 93.1126 87.8146 catagonus_wagneri ENSCWAP00000021069 93.1126 87.8146 ENSG00000169871 homo_sapiens ENSP00000305161 91.2583 87.5497 ictidomys_tridecemlineatus ENSSTOP00000018487 94.9036 91.0468 ENSG00000169871 homo_sapiens ENSP00000305161 90.4636 85.5629 otolemur_garnettii ENSOGAP00000019836 90.3439 85.4497 ENSG00000169871 homo_sapiens ENSP00000305161 96.8212 94.9669 cebus_imitator ENSCCAP00000001149 96.6931 94.8413 ENSG00000169871 homo_sapiens ENSP00000305161 72.9801 68.0795 cebus_imitator ENSCCAP00000018439 86.2285 80.4382 ENSG00000169871 homo_sapiens ENSP00000305161 93.245 89.1391 urocitellus_parryii ENSUPAP00010018342 88.8889 84.9747 ENSG00000169871 homo_sapiens ENSP00000305161 92.3179 87.5497 moschus_moschiferus ENSMMSP00000011538 92.3179 87.5497 ENSG00000169871 homo_sapiens ENSP00000305161 98.0132 96.5563 rhinopithecus_roxellana ENSRROP00000005795 98.0132 96.5563 ENSG00000169871 homo_sapiens ENSP00000305161 86.3576 80 mus_pahari MGP_PahariEiJ_P0059742 88.8283 82.2888 ENSG00000169871 homo_sapiens ENSP00000305161 34.3046 20.9272 ciona_intestinalis ENSCINP00000024518 30.6509 18.6982 ENSG00000169871 homo_sapiens ENSP00000305161 43.3113 23.5762 callorhinchus_milii ENSCMIP00000027867 48.2301 26.2537 ENSG00000169871 homo_sapiens ENSP00000305161 44.106 28.8742 lepisosteus_oculatus ENSLOCP00000022198 48.5423 31.7784 ENSG00000169871 homo_sapiens ENSP00000305161 33.3775 18.9404 clupea_harengus ENSCHAP00000012373 20.438 11.5977 ENSG00000169871 homo_sapiens ENSP00000305161 32.7152 16.4238 carassius_auratus ENSCARP00000125116 19.0586 9.5679 ENSG00000169871 homo_sapiens ENSP00000305161 31.1258 16.1589 astyanax_mexicanus ENSAMXP00000018856 18.5624 9.63665 ENSG00000169871 homo_sapiens ENSP00000305161 31.2583 16.5563 ictalurus_punctatus ENSIPUP00000006137 19.4719 10.3135 ENSG00000169871 homo_sapiens ENSP00000305161 33.5099 19.2053 ictalurus_punctatus ENSIPUP00000027478 25.4271 14.5729 ENSG00000169871 homo_sapiens ENSP00000305161 32.4503 18.4106 oncorhynchus_kisutch ENSOKIP00005033186 21.7006 12.3118 ENSG00000169871 homo_sapiens ENSP00000305161 32.3179 18.543 oncorhynchus_mykiss ENSOMYP00000050973 21.982 12.6126 ENSG00000169871 homo_sapiens ENSP00000305161 27.8146 15.6291 salmo_salar ENSSSAP00000102139 19.9052 11.1848 ENSG00000169871 homo_sapiens ENSP00000305161 31.6556 18.2781 salmo_trutta ENSSTUP00000040475 20.9833 12.1159 ENSG00000169871 homo_sapiens ENSP00000305161 26.2252 13.5099 oreochromis_niloticus ENSONIP00000071127 21.8785 11.2707 ENSG00000169871 homo_sapiens ENSP00000305161 27.8146 15.6291 haplochromis_burtoni ENSHBUP00000012216 14.7992 8.31571 ENSG00000169871 homo_sapiens ENSP00000305161 28.0795 15.6291 astatotilapia_calliptera ENSACLP00000018175 14.9401 8.31571 ENSG00000169871 homo_sapiens ENSP00000305161 24.106 13.3775 lates_calcarifer ENSLCAP00010028644 28.1734 15.6347 ENSG00000169871 homo_sapiens ENSP00000305161 36.9536 28.4768 chrysemys_picta_bellii ENSCPBP00000003284 55.0296 42.4063 ENSG00000169871 homo_sapiens ENSP00000305161 23.0464 17.351 sphenodon_punctatus ENSSPUP00000021584 67.7043 50.9728 ENSG00000169871 homo_sapiens ENSP00000305161 43.9735 27.0199 crocodylus_porosus ENSCPRP00005024912 48.4672 29.781 ENSG00000169871 homo_sapiens ENSP00000305161 56.2914 41.3245 laticauda_laticaudata ENSLLTP00000011784 62.592 45.9499 ENSG00000169871 homo_sapiens ENSP00000305161 55.7616 41.5894 notechis_scutatus ENSNSUP00000005503 62.1861 46.3811 ENSG00000169871 homo_sapiens ENSP00000305161 36.8212 27.4172 pseudonaja_textilis ENSPTXP00000022963 56.1616 41.8182 ENSG00000169871 homo_sapiens ENSP00000305161 24.5033 18.9404 anolis_carolinensis ENSACAP00000039863 69.5489 53.7594 ENSG00000169871 homo_sapiens ENSP00000305161 59.8675 44.2384 anolis_carolinensis ENSACAP00000007687 60.8345 44.9529 ENSG00000169871 homo_sapiens ENSP00000305161 11.1258 8.87417 naja_naja ENSNNAP00000015960 59.1549 47.1831 ENSG00000169871 homo_sapiens ENSP00000305161 61.8543 46.2252 podarcis_muralis ENSPMRP00000035890 60.3359 45.0904 ENSG00000169871 homo_sapiens ENSP00000305161 22.3841 17.4834 podarcis_muralis ENSPMRP00000035903 68.4211 53.4413 ENSG00000169871 homo_sapiens ENSP00000305161 60.2649 45.0331 podarcis_muralis ENSPMRP00000035864 61.7368 46.133 ENSG00000169871 homo_sapiens ENSP00000305161 40.7947 25.8278 erpetoichthys_calabaricus ENSECRP00000019772 42.5414 26.9337 ENSG00000169871 homo_sapiens ENSP00000305161 58.0132 44.5033 salvator_merianae ENSSMRP00000015437 63.0216 48.3453 ENSG00000169871 homo_sapiens ENSP00000305161 44.6358 33.5099 pelodiscus_sinensis ENSPSIP00000011096 57.6068 43.2479 ENSG00000169871 homo_sapiens ENSP00000305161 11.5232 9.27152 gopherus_evgoodei ENSGEVP00005024209 54.0373 43.4783 ENSG00000169871 homo_sapiens ENSP00000305161 22.3841 15.4967 chelonoidis_abingdonii ENSCABP00000025586 67.0635 46.4286 ENSG00000169871 homo_sapiens ENSP00000305161 13.6424 11.3907 chelonoidis_abingdonii ENSCABP00000020772 66.4516 55.4839 ENSG00000169871 homo_sapiens ENSP00000305161 43.5762 27.947 terrapene_carolina_triunguis ENSTMTP00000026532 48.1698 30.8931 ENSG00000169871 homo_sapiens ENSP00000305161 31.6556 17.8808 hucho_hucho ENSHHUP00000064270 21.4735 12.1294 ENSG00000169871 homo_sapiens ENSP00000305161 26.6225 13.6424 pygocentrus_nattereri ENSPNAP00000010698 17.8826 9.1637 ENSG00000169871 homo_sapiens ENSP00000305161 32.5828 18.543 ciona_savignyi ENSCSAVP00000007573 28.3084 16.1105 ENSG00000169871 homo_sapiens ENSP00000305161 32.3179 18.6755 oncorhynchus_tshawytscha ENSOTSP00005033303 21.6312 12.5 ENSG00000169871 homo_sapiens ENSP00000305161 42.2517 25.9603 paramormyrops_kingsleyae ENSPKIP00000024557 46.6374 28.655 ENSG00000169871 homo_sapiens ENSP00000305161 26.8874 18.4106 scleropages_formosus ENSSFOP00015071877 50.75 34.75
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0003723 enables RNA binding 1 HDA Homo_sapiens(9606) Function GO:0004842 enables ubiquitin-protein transferase activity 1 TAS Homo_sapiens(9606) Function GO:0005515 enables protein binding 1 IPI Homo_sapiens(9606) Function GO:0005737 located_in cytoplasm 1 ISS Homo_sapiens(9606) Component GO:0005829 located_in cytosol 1 TAS Homo_sapiens(9606) Component GO:0006513 involved_in protein monoubiquitination 2 IBA,IDA Homo_sapiens(9606) Process GO:0008270 enables zinc ion binding 1 IEA Homo_sapiens(9606) Function GO:0032479 involved_in regulation of type I interferon production 1 TAS Homo_sapiens(9606) Process GO:0032728 involved_in positive regulation of interferon-beta production 1 ISS Homo_sapiens(9606) Process GO:0034340 involved_in response to type I interferon 1 ISS Homo_sapiens(9606) Process GO:0045087 involved_in innate immune response 1 IBA Homo_sapiens(9606) Process GO:0045089 involved_in positive regulation of innate immune response 1 IDA Homo_sapiens(9606) Process GO:0051607 involved_in defense response to virus 2 IDA,ISS Homo_sapiens(9606) Process GO:0060340 involved_in positive regulation of type I interferon-mediated signaling pathway 2 IBA,IDA Homo_sapiens(9606) Process GO:0061630 enables ubiquitin protein ligase activity 3 IBA,IDA,ISS Homo_sapiens(9606) Function GO:0070534 involved_in protein K63-linked ubiquitination 1 ISS Homo_sapiens(9606) Process GO:1901224 involved_in positive regulation of NIK/NF-kappaB signaling 1 IDA Homo_sapiens(9606) Process
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.