RBP Type
Non-canonical_RBPs
Diseases
SARS-COV-2
Drug
N.A.
Main interacting RNAs
N.A.
Moonlighting functions
Transmemebrane
Localizations
P-body
BulkPerturb-seq
N.A.
Ensembl ID ENSG00000169100 Gene ID
293 Accession
10992
Symbol
SLC25A6
Alias
ANT;AAC3;ANT3;ANT2;ANT3;ANT3Y
Full Name
solute carrier family 25 member 6
Status
Confidence
Length
5962 bases
Strand
Minus strand
Position
X : 1386152 - 1392113
RNA binding domain
N.A.
Summary
This gene is a member of the mitochondrial carrier subfamily of solute carrier protein genes. The product of this gene functions as a gated pore that translocates ADP from the cytoplasm into the mitochondrial matrix and ATP from the mitochondrial matrix into the cytoplasm. The protein is implicated in the function of the permability transition pore complex (PTPC), which regulates the release of mitochondrial products that induce apoptosis. The human genome contains several non-transcribed pseudogenes of this gene. [provided by RefSeq, Jun 2013]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
Name
Transcript ID
bp
Protein
Translation ID
SLC25A6-201
ENST00000381401
1451
298aa
ENSP00000370808
SLC25A6-202
ENST00000475167
900
No protein
-
SLC25A6-203
ENST00000484026
854
No protein
-
Pathway ID
Pathway Name
Source
ensgID
Trait
pValue
Pubmed ID
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
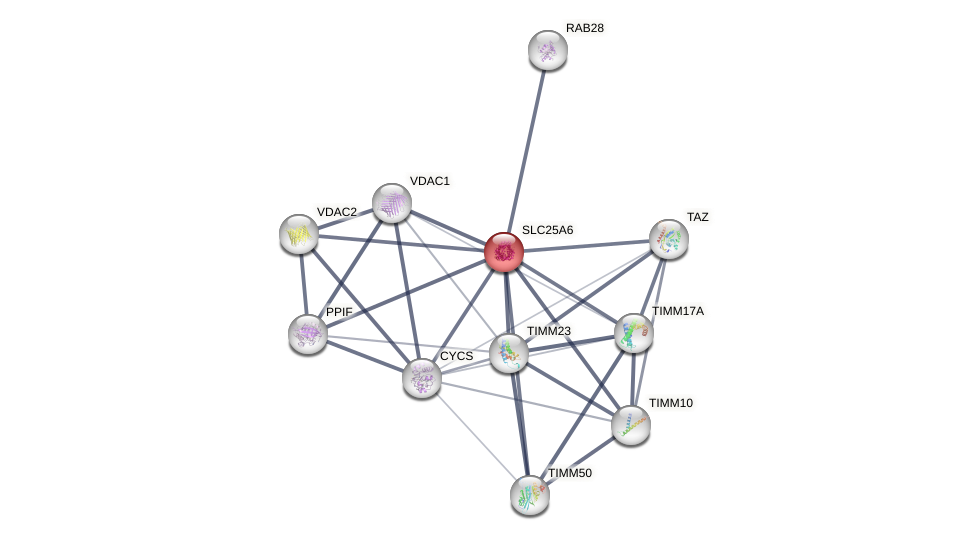
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
ENSG00000169100 homo_sapiens ENSP00000464231 34.6667 16.6667 homo_sapiens ENSP00000370808 34.8993 16.7785 ENSG00000169100 homo_sapiens ENSP00000320688 38.5852 22.508 homo_sapiens ENSP00000370808 40.2685 23.4899 ENSG00000169100 homo_sapiens ENSP00000326693 41.8239 25.7862 homo_sapiens ENSP00000370808 44.6309 27.5168 ENSG00000169100 homo_sapiens ENSP00000242275 35.0168 19.5286 homo_sapiens ENSP00000370808 34.8993 19.4631 ENSG00000169100 homo_sapiens ENSP00000372612 35.0168 18.8552 homo_sapiens ENSP00000370808 34.8993 18.7919 ENSG00000169100 homo_sapiens ENSP00000497532 34.5395 19.0789 homo_sapiens ENSP00000370808 35.2349 19.4631 ENSG00000169100 homo_sapiens ENSP00000217909 41.9355 25.5132 homo_sapiens ENSP00000370808 47.9866 29.1946 ENSG00000169100 homo_sapiens ENSP00000429200 33.432 16.2722 homo_sapiens ENSP00000370808 37.9195 18.4564 ENSG00000169100 homo_sapiens ENSP00000429168 39.5189 24.055 homo_sapiens ENSP00000370808 38.5906 23.4899 ENSG00000169100 homo_sapiens ENSP00000344831 31.3609 17.4556 homo_sapiens ENSP00000370808 35.5705 19.7987 ENSG00000169100 homo_sapiens ENSP00000354886 35.7143 21.7532 homo_sapiens ENSP00000370808 36.9128 22.4832 ENSG00000169100 homo_sapiens ENSP00000322649 38.9189 21.0811 homo_sapiens ENSP00000370808 48.3221 26.1745 ENSG00000169100 homo_sapiens ENSP00000329452 42.1405 24.4147 homo_sapiens ENSP00000370808 42.2819 24.4966 ENSG00000169100 homo_sapiens ENSP00000457733 33.3333 19.7065 homo_sapiens ENSP00000370808 53.3557 31.5436 ENSG00000169100 homo_sapiens ENSP00000359526 31.3187 17.3077 homo_sapiens ENSP00000370808 38.255 21.1409 ENSG00000169100 homo_sapiens ENSP00000360671 96.3087 91.9463 homo_sapiens ENSP00000370808 96.3087 91.9463 ENSG00000169100 homo_sapiens ENSP00000345580 36.5854 20.9059 homo_sapiens ENSP00000370808 35.2349 20.1342 ENSG00000169100 homo_sapiens ENSP00000360398 31.8885 18.5759 homo_sapiens ENSP00000370808 34.5638 20.1342 ENSG00000169100 homo_sapiens ENSP00000326305 37.2093 23.588 homo_sapiens ENSP00000370808 37.5839 23.8255 ENSG00000169100 homo_sapiens ENSP00000448708 25.4848 12.1884 homo_sapiens ENSP00000370808 30.8725 14.7651 ENSG00000169100 homo_sapiens ENSP00000397818 35.625 20.625 homo_sapiens ENSP00000370808 38.255 22.1477 ENSG00000169100 homo_sapiens ENSP00000388658 20.5015 13.2743 homo_sapiens ENSP00000370808 46.6443 30.2013 ENSG00000169100 homo_sapiens ENSP00000486058 34.3653 21.3622 homo_sapiens ENSP00000370808 37.2483 23.1544 ENSG00000169100 homo_sapiens ENSP00000329033 37.1429 20 homo_sapiens ENSP00000370808 39.2617 21.1409 ENSG00000169100 homo_sapiens ENSP00000262999 33.8762 20.8469 homo_sapiens ENSP00000370808 34.8993 21.4765 ENSG00000169100 homo_sapiens ENSP00000323740 35.2564 22.4359 homo_sapiens ENSP00000370808 36.9128 23.4899 ENSG00000169100 homo_sapiens ENSP00000225665 32.1656 17.5159 homo_sapiens ENSP00000370808 33.8926 18.4564 ENSG00000169100 homo_sapiens ENSP00000476815 38.253 24.3976 homo_sapiens ENSP00000370808 42.6175 27.1812 ENSG00000169100 homo_sapiens ENSP00000306328 36.4486 20.8723 homo_sapiens ENSP00000370808 39.2617 22.4832 ENSG00000169100 homo_sapiens ENSP00000362160 29.3204 16.8932 homo_sapiens ENSP00000370808 50.6711 29.1946 ENSG00000169100 homo_sapiens ENSP00000281154 78.4127 66.6667 homo_sapiens ENSP00000370808 82.8859 70.4698 ENSG00000169100 homo_sapiens ENSP00000352497 27.3885 15.9236 homo_sapiens ENSP00000370808 28.8591 16.7785 ENSG00000169100 homo_sapiens ENSP00000215882 36.9775 22.508 homo_sapiens ENSP00000370808 38.5906 23.4899 ENSG00000169100 homo_sapiens ENSP00000366299 28.1337 17.2702 homo_sapiens ENSP00000370808 33.8926 20.8054 ENSG00000169100 homo_sapiens ENSP00000499695 35.2751 21.3592 homo_sapiens ENSP00000370808 36.5772 22.1477 ENSG00000169100 homo_sapiens ENSP00000342267 35.2159 18.2724 homo_sapiens ENSP00000370808 35.5705 18.4564 ENSG00000169100 homo_sapiens ENSP00000468980 23.4528 12.0521 homo_sapiens ENSP00000370808 24.1611 12.4161 ENSG00000169100 homo_sapiens ENSP00000381782 39.2361 24.3056 homo_sapiens ENSP00000370808 37.9195 23.4899 ENSG00000169100 homo_sapiens ENSP00000506858 37.6206 20.9003 homo_sapiens ENSP00000370808 39.2617 21.8121 ENSG00000169100 homo_sapiens ENSP00000301454 33.1197 18.8034 homo_sapiens ENSP00000370808 52.0134 29.5302 ENSG00000169100 homo_sapiens ENSP00000239451 35.8804 19.2691 homo_sapiens ENSP00000370808 36.2416 19.4631 ENSG00000169100 homo_sapiens ENSP00000346955 28.8321 14.5985 homo_sapiens ENSP00000370808 26.5101 13.4228 ENSG00000169100 homo_sapiens ENSP00000352167 42.5743 26.0726 homo_sapiens ENSP00000370808 43.2886 26.5101 ENSG00000169100 homo_sapiens ENSP00000390722 29.9674 18.241 homo_sapiens ENSP00000370808 30.8725 18.7919 ENSG00000169100 homo_sapiens ENSP00000281456 94.2953 87.9195 homo_sapiens ENSP00000370808 94.2953 87.9195 ENSG00000169100 homo_sapiens ENSP00000294454 33.8816 19.0789 homo_sapiens ENSP00000370808 34.5638 19.4631 ENSG00000169100 homo_sapiens ENSP00000297578 40 24.7619 homo_sapiens ENSP00000370808 42.2819 26.1745 ENSG00000169100 homo_sapiens ENSP00000265631 19.7037 12.2963 homo_sapiens ENSP00000370808 44.6309 27.8523 ENSG00000169100 homo_sapiens ENSP00000444642 34.4615 21.5385 homo_sapiens ENSP00000370808 37.5839 23.4899
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
ENSG00000169100 homo_sapiens ENSP00000370808 99.3289 98.9933 nomascus_leucogenys ENSNLEP00000013529 99.3289 98.9933 ENSG00000169100 homo_sapiens ENSP00000370808 100 100 pongo_abelii ENSPPYP00000037639 100 100 ENSG00000169100 homo_sapiens ENSP00000370808 100 100 pan_troglodytes ENSPTRP00000063171 100 100 ENSG00000169100 homo_sapiens ENSP00000370808 100 100 gorilla_gorilla ENSGGOP00000027561 100 100 ENSG00000169100 homo_sapiens ENSP00000370808 53.3557 48.9933 erinaceus_europaeus ENSEEUP00000002245 87.8453 80.663 ENSG00000169100 homo_sapiens ENSP00000370808 98.9933 94.9664 echinops_telfairi ENSETEP00000005698 98.9933 94.9664 ENSG00000169100 homo_sapiens ENSP00000370808 22.8188 21.1409 echinops_telfairi ENSETEP00000004329 94.4444 87.5 ENSG00000169100 homo_sapiens ENSP00000370808 100 99.6644 callithrix_jacchus ENSCJAP00000066084 100 99.6644 ENSG00000169100 homo_sapiens ENSP00000370808 68.4564 66.7785 callithrix_jacchus ENSCJAP00000062305 94.8837 92.5581 ENSG00000169100 homo_sapiens ENSP00000370808 100 99.6644 callithrix_jacchus ENSCJAP00000079481 100 99.6644 ENSG00000169100 homo_sapiens ENSP00000370808 100 99.6644 cercocebus_atys ENSCATP00000002634 100 99.6644 ENSG00000169100 homo_sapiens ENSP00000370808 93.6242 92.2819 macaca_fascicularis ENSMFAP00000023608 95.5479 94.1781 ENSG00000169100 homo_sapiens ENSP00000370808 89.5973 86.5772 macaca_mulatta ENSMMUP00000063020 66.9173 64.6617 ENSG00000169100 homo_sapiens ENSP00000370808 100 100 macaca_nemestrina ENSMNEP00000020654 100 100 ENSG00000169100 homo_sapiens ENSP00000370808 99.6644 97.9866 tursiops_truncatus ENSTTRP00000014594 99.6644 97.9866 ENSG00000169100 homo_sapiens ENSP00000370808 99.3289 97.9866 capra_hircus ENSCHIP00000009218 99.3289 97.9866 ENSG00000169100 homo_sapiens ENSP00000370808 99.6644 96.6443 loxodonta_africana ENSLAFP00000001322 99.6644 96.6443 ENSG00000169100 homo_sapiens ENSP00000370808 99.6644 97.9866 panthera_leo ENSPLOP00000022301 99.6644 97.9866 ENSG00000169100 homo_sapiens ENSP00000370808 97.3154 91.6107 ailuropoda_melanoleuca ENSAMEP00000032567 97.3154 91.6107 ENSG00000169100 homo_sapiens ENSP00000370808 97.9866 92.6174 ailuropoda_melanoleuca ENSAMEP00000042609 97.9866 92.6174 ENSG00000169100 homo_sapiens ENSP00000370808 68.1208 64.4295 ailuropoda_melanoleuca ENSAMEP00000033015 97.5962 92.3077 ENSG00000169100 homo_sapiens ENSP00000370808 99.6644 98.6577 canis_lupus_familiaris ENSCAFP00845030136 99.6644 98.6577 ENSG00000169100 homo_sapiens ENSP00000370808 99.3289 97.3154 balaenoptera_musculus ENSBMSP00010020498 99.3289 97.3154 ENSG00000169100 homo_sapiens ENSP00000370808 98.6577 96.6443 felis_catus ENSFCAP00000048393 92.4528 90.566 ENSG00000169100 homo_sapiens ENSP00000370808 71.4765 66.443 monodelphis_domestica ENSMODP00000026274 93.8326 87.2247 ENSG00000169100 homo_sapiens ENSP00000370808 99.6644 96.3087 monodelphis_domestica ENSMODP00000036397 99.6644 96.3087 ENSG00000169100 homo_sapiens ENSP00000370808 99.6644 96.6443 monodelphis_domestica ENSMODP00000002634 99.6644 96.6443 ENSG00000169100 homo_sapiens ENSP00000370808 99.6644 97.651 monodon_monoceros ENSMMNP00015025158 99.6644 97.651 ENSG00000169100 homo_sapiens ENSP00000370808 99.3289 97.651 sus_scrofa ENSSSCP00000072750 99.3289 97.651 ENSG00000169100 homo_sapiens ENSP00000370808 99.6644 98.6577 microcebus_murinus ENSMICP00000002505 99.6644 98.6577 ENSG00000169100 homo_sapiens ENSP00000370808 99.6644 97.9866 delphinapterus_leucas ENSDLEP00000026268 99.6644 97.9866 ENSG00000169100 homo_sapiens ENSP00000370808 99.6644 97.651 physeter_catodon ENSPCTP00005014160 99.6644 97.651 ENSG00000169100 homo_sapiens ENSP00000370808 91.2752 90.604 aotus_nancymaae ENSANAP00000002681 98.9091 98.1818 ENSG00000169100 homo_sapiens ENSP00000370808 92.2819 89.5973 aotus_nancymaae ENSANAP00000022025 94.8276 92.069 ENSG00000169100 homo_sapiens ENSP00000370808 92.2819 89.5973 aotus_nancymaae ENSANAP00000013941 94.8276 92.069 ENSG00000169100 homo_sapiens ENSP00000370808 98.3222 97.651 saimiri_boliviensis_boliviensis ENSSBOP00000002452 98.3222 97.651 ENSG00000169100 homo_sapiens ENSP00000370808 51.3423 51.3423 saimiri_boliviensis_boliviensis ENSSBOP00000033607 98.0769 98.0769 ENSG00000169100 homo_sapiens ENSP00000370808 100 100 chlorocebus_sabaeus ENSCSAP00000014089 100 100 ENSG00000169100 homo_sapiens ENSP00000370808 60.7383 56.3758 camelus_dromedarius ENSCDRP00005025462 85.782 79.6208 ENSG00000169100 homo_sapiens ENSP00000370808 99.6644 98.6577 prolemur_simus ENSPSMP00000037589 99.6644 98.6577 ENSG00000169100 homo_sapiens ENSP00000370808 45.9732 41.6107 sorex_araneus ENSSARP00000001290 88.3871 80 ENSG00000169100 homo_sapiens ENSP00000370808 99.6644 96.9799 vombatus_ursinus ENSVURP00010020474 99.6644 96.9799 ENSG00000169100 homo_sapiens ENSP00000370808 99.3289 96.6443 rhinolophus_ferrumequinum ENSRFEP00010006670 99.3289 96.6443 ENSG00000169100 homo_sapiens ENSP00000370808 100 100 rhinopithecus_bieti ENSRBIP00000026202 100 100 ENSG00000169100 homo_sapiens ENSP00000370808 98.3222 97.3154 canis_lupus_dingo ENSCAFP00020001568 92.7215 91.7721 ENSG00000169100 homo_sapiens ENSP00000370808 99.3289 97.651 sciurus_vulgaris ENSSVLP00005031510 99.3289 97.651 ENSG00000169100 homo_sapiens ENSP00000370808 99.3289 97.3154 phascolarctos_cinereus ENSPCIP00000010199 99.3289 97.3154 ENSG00000169100 homo_sapiens ENSP00000370808 79.1946 74.8322 procavia_capensis ENSPCAP00000007102 95.935 90.6504 ENSG00000169100 homo_sapiens ENSP00000370808 99.6644 94.2953 nannospalax_galili ENSNGAP00000012430 99.6644 94.2953 ENSG00000169100 homo_sapiens ENSP00000370808 89.9329 85.2349 bos_grunniens ENSBGRP00000011461 75.9207 71.9547 ENSG00000169100 homo_sapiens ENSP00000370808 99.6644 97.9866 catagonus_wagneri ENSCWAP00000026902 99.6644 97.9866 ENSG00000169100 homo_sapiens ENSP00000370808 99.3289 98.9933 cebus_imitator ENSCCAP00000015729 99.3289 98.9933 ENSG00000169100 homo_sapiens ENSP00000370808 99.3289 97.3154 moschus_moschiferus ENSMMSP00000022429 99.3289 97.3154 ENSG00000169100 homo_sapiens ENSP00000370808 100 100 rhinopithecus_roxellana ENSRROP00000028957 100 100 ENSG00000169100 homo_sapiens ENSP00000370808 99.3289 98.9933 propithecus_coquereli ENSPCOP00000022040 99.3289 98.9933 ENSG00000169100 homo_sapiens ENSP00000370808 95.6376 90.9396 latimeria_chalumnae ENSLACP00000010337 95.6376 90.9396 ENSG00000169100 homo_sapiens ENSP00000370808 83.2215 76.8456 clupea_harengus ENSCHAP00000044216 74.0299 68.3582 ENSG00000169100 homo_sapiens ENSP00000370808 94.9664 91.9463 danio_rerio ENSDARP00000022077 94.9664 91.9463 ENSG00000169100 homo_sapiens ENSP00000370808 66.7785 63.7584 carassius_auratus ENSCARP00000080253 85.0427 81.1966 ENSG00000169100 homo_sapiens ENSP00000370808 44.2953 41.2752 carassius_auratus ENSCARP00000008079 96.3504 89.781 ENSG00000169100 homo_sapiens ENSP00000370808 91.6107 85.906 carassius_auratus ENSCARP00000092072 90.099 84.4884 ENSG00000169100 homo_sapiens ENSP00000370808 85.5705 76.5101 astyanax_mexicanus ENSAMXP00000044431 86.4407 77.2881 ENSG00000169100 homo_sapiens ENSP00000370808 90.2685 83.8926 astyanax_mexicanus ENSAMXP00000041144 89.6667 83.3333 ENSG00000169100 homo_sapiens ENSP00000370808 93.6242 88.5906 ictalurus_punctatus ENSIPUP00000034879 93.311 88.2943 ENSG00000169100 homo_sapiens ENSP00000370808 93.6242 89.2617 esox_lucius ENSELUP00000010514 93.311 88.9632 ENSG00000169100 homo_sapiens ENSP00000370808 94.9664 89.5973 electrophorus_electricus ENSEEEP00000047312 90.7051 85.5769 ENSG00000169100 homo_sapiens ENSP00000370808 83.2215 73.1544 oncorhynchus_kisutch ENSOKIP00005113507 81.5789 71.7105 ENSG00000169100 homo_sapiens ENSP00000370808 82.5503 76.1745 salmo_salar ENSSSAP00000056860 82.5503 76.1745 ENSG00000169100 homo_sapiens ENSP00000370808 92.6174 87.2483 gadus_morhua ENSGMOP00000021110 89.6104 84.4156 ENSG00000169100 homo_sapiens ENSP00000370808 26.5101 24.4966 fundulus_heteroclitus ENSFHEP00000023300 77.451 71.5686 ENSG00000169100 homo_sapiens ENSP00000370808 93.6242 87.9195 fundulus_heteroclitus ENSFHEP00000013998 93.311 87.6254 ENSG00000169100 homo_sapiens ENSP00000370808 95.9732 92.2819 fundulus_heteroclitus ENSFHEP00000014036 95.6522 91.9732 ENSG00000169100 homo_sapiens ENSP00000370808 93.6242 86.5772 poecilia_reticulata ENSPREP00000013235 93.311 86.2876 ENSG00000169100 homo_sapiens ENSP00000370808 96.9799 93.2886 poecilia_reticulata ENSPREP00000013259 78.3198 75.3388 ENSG00000169100 homo_sapiens ENSP00000370808 96.9799 92.953 xiphophorus_maculatus ENSXMAP00000012936 96.6555 92.6421 ENSG00000169100 homo_sapiens ENSP00000370808 93.6242 87.2483 xiphophorus_maculatus ENSXMAP00000022258 93.9394 87.5421 ENSG00000169100 homo_sapiens ENSP00000370808 95.9732 91.6107 oryzias_latipes ENSORLP00000030127 95.9732 91.6107 ENSG00000169100 homo_sapiens ENSP00000370808 88.9262 82.5503 cyclopterus_lumpus ENSCLMP00005026385 88.6288 82.2742 ENSG00000169100 homo_sapiens ENSP00000370808 93.9597 88.9262 cyclopterus_lumpus ENSCLMP00005029538 89.4569 84.6645 ENSG00000169100 homo_sapiens ENSP00000370808 90.2685 82.8859 oreochromis_niloticus ENSONIP00000075297 89.9666 82.6087 ENSG00000169100 homo_sapiens ENSP00000370808 95.302 90.2685 sparus_aurata ENSSAUP00010066945 92.8105 87.9085 ENSG00000169100 homo_sapiens ENSP00000370808 91.2752 84.5638 lates_calcarifer ENSLCAP00010014022 90.6667 84 ENSG00000169100 homo_sapiens ENSP00000370808 97.3154 89.9329 notechis_scutatus ENSNSUP00000009554 97.3154 89.9329 ENSG00000169100 homo_sapiens ENSP00000370808 65.7718 58.3893 pseudonaja_textilis ENSPTXP00000021151 89.0909 79.0909 ENSG00000169100 homo_sapiens ENSP00000370808 97.9866 93.6242 anas_platyrhynchos_platyrhynchos ENSAPLP00000029563 88.2175 84.29 ENSG00000169100 homo_sapiens ENSP00000370808 97.9866 93.2886 parus_major ENSPMJP00000000597 97.9866 93.2886 ENSG00000169100 homo_sapiens ENSP00000370808 96.9799 91.9463 naja_naja ENSNNAP00000019409 96.9799 91.9463 ENSG00000169100 homo_sapiens ENSP00000370808 81.2081 75.5034 podarcis_muralis ENSPMRP00000030501 78.0645 72.5806 ENSG00000169100 homo_sapiens ENSP00000370808 97.651 91.9463 salvator_merianae ENSSMRP00000029165 97.651 91.9463 ENSG00000169100 homo_sapiens ENSP00000370808 97.3154 93.2886 amphilophus_citrinellus ENSACIP00000013504 96.99 92.9766 ENSG00000169100 homo_sapiens ENSP00000370808 94.2953 86.5772 amphilophus_citrinellus ENSACIP00000013522 94.2953 86.5772 ENSG00000169100 homo_sapiens ENSP00000370808 90.9396 84.8993 dicentrarchus_labrax ENSDLAP00005046286 88.2736 82.4104 ENSG00000169100 homo_sapiens ENSP00000370808 83.8926 76.1745 dicentrarchus_labrax ENSDLAP00005008828 80.6452 73.2258 ENSG00000169100 homo_sapiens ENSP00000370808 98.9933 92.953 gopherus_evgoodei ENSGEVP00005005096 98.9933 92.953 ENSG00000169100 homo_sapiens ENSP00000370808 98.9933 92.953 chelonoidis_abingdonii ENSCABP00000031513 98.9933 92.953 ENSG00000169100 homo_sapiens ENSP00000370808 90.2685 84.5638 cottoperca_gobio ENSCGOP00000054354 89.6667 84 ENSG00000169100 homo_sapiens ENSP00000370808 97.9866 93.6242 anser_brachyrhynchus ENSABRP00000021143 97.9866 93.6242 ENSG00000169100 homo_sapiens ENSP00000370808 92.2819 85.2349 cynoglossus_semilaevis ENSCSEP00000000315 90.4605 83.5526 ENSG00000169100 homo_sapiens ENSP00000370808 96.9799 93.6242 poecilia_formosa ENSPFOP00000009667 96.9799 93.6242 ENSG00000169100 homo_sapiens ENSP00000370808 77.5168 73.4899 poecilia_formosa ENSPFOP00000006290 92.0319 87.251 ENSG00000169100 homo_sapiens ENSP00000370808 94.2953 91.2752 myripristis_murdjan ENSMMDP00005045657 94.2953 91.2752 ENSG00000169100 homo_sapiens ENSP00000370808 87.2483 80.8725 sander_lucioperca ENSSLUP00000030051 86.3787 80.0664 ENSG00000169100 homo_sapiens ENSP00000370808 97.9866 93.2886 strigops_habroptila ENSSHBP00005012013 97.9866 93.2886 ENSG00000169100 homo_sapiens ENSP00000370808 98.9933 92.953 terrapene_carolina_triunguis ENSTMTP00000008387 98.9933 92.953 ENSG00000169100 homo_sapiens ENSP00000370808 98.9933 92.953 terrapene_carolina_triunguis ENSTMTP00000014952 98.9933 92.953 ENSG00000169100 homo_sapiens ENSP00000370808 95.6376 91.9463 pygocentrus_nattereri ENSPNAP00000031945 95.6376 91.9463 ENSG00000169100 homo_sapiens ENSP00000370808 97.3154 92.6174 labrus_bergylta ENSLBEP00000009936 97.3154 92.6174 ENSG00000169100 homo_sapiens ENSP00000370808 91.6107 85.2349 mastacembelus_armatus ENSMAMP00000026606 89.2157 83.0065 ENSG00000169100 homo_sapiens ENSP00000370808 87.5839 79.8658 oncorhynchus_tshawytscha ENSOTSP00005020819 87.8788 80.1347 ENSG00000169100 homo_sapiens ENSP00000370808 91.9463 86.9128 aquila_chrysaetos_chrysaetos ENSACCP00020012370 88.1029 83.2797 ENSG00000169100 homo_sapiens ENSP00000370808 96.6443 91.9463 nothobranchius_furzeri ENSNFUP00015003176 96.6443 91.9463 ENSG00000169100 homo_sapiens ENSP00000370808 96.6443 92.2819 cyprinodon_variegatus ENSCVAP00000015374 96.3211 91.9732 ENSG00000169100 homo_sapiens ENSP00000370808 95.6376 91.2752 denticeps_clupeoides ENSDCDP00000061490 80.5085 76.8362 ENSG00000169100 homo_sapiens ENSP00000370808 95.6376 91.6107 larimichthys_crocea ENSLCRP00005050709 88.785 85.0467 ENSG00000169100 homo_sapiens ENSP00000370808 86.9128 81.8792 ficedula_albicollis ENSFALP00000001667 85.7616 80.7947 ENSG00000169100 homo_sapiens ENSP00000370808 87.2483 78.5235 cyprinus_carpio_carpio ENSCCRP00000135254 70.4607 63.4146 ENSG00000169100 homo_sapiens ENSP00000370808 97.9866 93.6242 coturnix_japonica ENSCJPP00005022969 97.9866 93.6242 ENSG00000169100 homo_sapiens ENSP00000370808 96.9799 93.6242 poecilia_latipinna ENSPLAP00000008616 96.6555 93.311 ENSG00000169100 homo_sapiens ENSP00000370808 95.302 90.2685 sinocyclocheilus_grahami ENSSGRP00000082395 93.4211 88.4868 ENSG00000169100 homo_sapiens ENSP00000370808 94.6309 89.9329 sinocyclocheilus_grahami ENSSGRP00000066700 85.9756 81.7073 ENSG00000169100 homo_sapiens ENSP00000370808 90.9396 84.5638 paramormyrops_kingsleyae ENSPKIP00000020393 90.9396 84.5638 ENSG00000169100 homo_sapiens ENSP00000370808 89.5973 84.5638 scleropages_formosus ENSSFOP00015034594 85.5769 80.7692 ENSG00000169100 homo_sapiens ENSP00000370808 94.9664 89.9329 leptobrachium_leishanense ENSLLEP00000045550 94.9664 89.9329 ENSG00000169100 homo_sapiens ENSP00000370808 96.6443 91.6107 scophthalmus_maximus ENSSMAP00000026731 90.2821 85.5799
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0005471 enables ATP:ADP antiporter activity 1 NAS Homo_sapiens(9606) Function GO:0005515 enables protein binding 1 IPI Homo_sapiens(9606) Function GO:0005634 located_in nucleus 1 HDA Homo_sapiens(9606) Component GO:0005739 located_in mitochondrion 2 HDA,IDA Homo_sapiens(9606) Component GO:0005743 located_in mitochondrial inner membrane 1 TAS Homo_sapiens(9606) Component GO:0005744 part_of TIM23 mitochondrial import inner membrane translocase complex 1 TAS Homo_sapiens(9606) Component GO:0006915 involved_in apoptotic process 1 IEA Homo_sapiens(9606) Process GO:0016020 located_in membrane 1 IDA Homo_sapiens(9606) Component GO:0140021 involved_in mitochondrial ADP transmembrane transport 1 IEA Homo_sapiens(9606) Process GO:1901029 involved_in negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway 1 IBA Homo_sapiens(9606) Process GO:1990544 involved_in mitochondrial ATP transmembrane transport 1 IEA Homo_sapiens(9606) Process
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.