Welcome to
RBP World!
Description
| Ensembl ID | ENSG00000168610 | Gene ID | 6774 | Accession | 11364 |
| Symbol | STAT3 | Alias | APRF;HIES;ADMIO;ADMIO1 | Full Name | signal transducer and activator of transcription 3 |
| Status | Confidence | Length | 75217 bases | Strand | Minus strand |
| Position | 17 : 42313324 - 42388540 | RNA binding domain | N.A. | ||
| Summary | The protein encoded by this gene is a member of the STAT protein family. In response to cytokines and growth factors, STAT family members are phosphorylated by the receptor associated kinases, and then form homo- or heterodimers that translocate to the cell nucleus where they act as transcription activators. This protein is activated through phosphorylation in response to various cytokines and growth factors including IFNs, EGF, IL5, IL6, HGF, LIF and BMP2. This protein mediates the expression of a variety of genes in response to cell stimuli, and thus plays a key role in many cellular processes such as cell growth and apoptosis. The small GTPase Rac1 has been shown to bind and regulate the activity of this protein. PIAS3 protein is a specific inhibitor of this protein. This gene also plays a role in regulating host response to viral and bacterial infections. Mutations in this gene are associated with infantile-onset multisystem autoimmune disease and hyper-immunoglobulin E syndrome. [provided by RefSeq, Aug 2020] | ||||
RNA binding domains (RBDs)
| Protein | Domain | Pfam ID | E-value | Domain number |
|---|
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|---|---|---|---|
| 36516770 | Splicing factor deficits render hematopoietic stem and progenitor cells sensitive to STAT3 inhibition. | S Kathryn Potts | 2022-12-13 | Cell reports |
| 18503751 | An RNA biding protein, Y14 interacts with and modulates STAT3 activation. | Norihiko Ohbayashi | 2008-08-01 | Biochemical and biophysical research communications |
| 29914167 | STAT3 Interactors as Potential Therapeutic Targets for Cancer Treatment. | Federica Laudisi | 2018-06-16 | International journal of molecular sciences |
| 33225311 | Genomic characterization of HIV-associated plasmablastic lymphoma identifies pervasive mutations in the JAK-STAT pathway. | Zhaoqi Liu | 2020-07-01 | Blood cancer discovery |
| 27486347 | Oncogenes: The Passport for Viral Oncolysis Through PKR Inhibition. | Janaina Fernandes | 2016-01-01 | Biomarkers in cancer |
| 35565466 | Oncopeptide MBOP Encoded by LINC01234 Promotes Colorectal Cancer through MAPK Signaling Pathway. | Chunyuan Tang | 2022-05-09 | Cancers |
| 31988283 | LncRNA AC093818.1 accelerates gastric cancer metastasis by epigenetically promoting PDK1 expression. | Ming-Chen Ba | 2020-01-27 | Cell death & disease |
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| STAT3-201 | ENST00000264657 | 4921 | 770aa | ENSP00000264657 |
| STAT3-246 | ENST00000678960 | 4912 | 770aa | ENSP00000503181 |
| STAT3-244 | ENST00000678906 | 4971 | 770aa | ENSP00000504184 |
| STAT3-231 | ENST00000678044 | 4912 | 770aa | ENSP00000503102 |
| STAT3-250 | ENST00000679231 | 4880 | No protein | - |
| STAT3-222 | ENST00000677421 | 4911 | 802aa | ENSP00000503599 |
| STAT3-232 | ENST00000678048 | 4740 | 742aa | ENSP00000503799 |
| STAT3-239 | ENST00000678674 | 4703 | 737aa | ENSP00000504062 |
| STAT3-234 | ENST00000678445 | 4944 | 575aa | ENSP00000503105 |
| STAT3-245 | ENST00000678913 | 4839 | 775aa | ENSP00000504609 |
| STAT3-240 | ENST00000678764 | 5054 | No protein | - |
| STAT3-241 | ENST00000678792 | 4842 | 775aa | ENSP00000504435 |
| STAT3-248 | ENST00000679166 | 4748 | 742aa | ENSP00000503308 |
| STAT3-223 | ENST00000677442 | 4829 | 761aa | ENSP00000504350 |
| STAT3-227 | ENST00000677723 | 4834 | 769aa | ENSP00000503574 |
| STAT3-230 | ENST00000678043 | 4837 | 763aa | ENSP00000503872 |
| STAT3-247 | ENST00000679014 | 4890 | 769aa | ENSP00000503237 |
| STAT3-216 | ENST00000677030 | 4812 | 722aa | ENSP00000503662 |
| STAT3-214 | ENST00000676636 | 4822 | 575aa | ENSP00000504255 |
| STAT3-237 | ENST00000678572 | 4739 | 727aa | ENSP00000504182 |
| STAT3-219 | ENST00000677271 | 4489 | 458aa | ENSP00000503912 |
| STAT3-243 | ENST00000678905 | 4768 | 726aa | ENSP00000503333 |
| STAT3-224 | ENST00000677479 | 4731 | 721aa | ENSP00000503559 |
| STAT3-229 | ENST00000677820 | 4879 | 575aa | ENSP00000504715 |
| STAT3-249 | ENST00000679185 | 4782 | 721aa | ENSP00000503332 |
| STAT3-242 | ENST00000678827 | 4800 | 722aa | ENSP00000503634 |
| STAT3-205 | ENST00000462286 | 1248 | No protein | - |
| STAT3-221 | ENST00000677346 | 1486 | No protein | - |
| STAT3-208 | ENST00000491272 | 1327 | No protein | - |
| STAT3-225 | ENST00000677500 | 1277 | No protein | - |
| STAT3-215 | ENST00000677002 | 3180 | 760aa | ENSP00000503742 |
| STAT3-217 | ENST00000677152 | 3395 | 765aa | ENSP00000502874 |
| STAT3-226 | ENST00000677603 | 3314 | 738aa | ENSP00000504324 |
| STAT3-211 | ENST00000585517 | 3319 | 722aa | ENSP00000467000 |
| STAT3-220 | ENST00000677308 | 3374 | 575aa | ENSP00000503059 |
| STAT3-236 | ENST00000678535 | 3288 | 738aa | ENSP00000504081 |
| STAT3-202 | ENST00000389272 | 2615 | 672aa | ENSP00000373923 |
| STAT3-212 | ENST00000588969 | 2772 | 770aa | ENSP00000467985 |
| STAT3-228 | ENST00000677763 | 3027 | No protein | - |
| STAT3-203 | ENST00000404395 | 2633 | 769aa | ENSP00000384943 |
| STAT3-204 | ENST00000462269 | 698 | No protein | - |
| STAT3-218 | ENST00000677270 | 2680 | No protein | - |
| STAT3-238 | ENST00000678659 | 2866 | No protein | - |
| STAT3-206 | ENST00000471989 | 826 | No protein | - |
| STAT3-209 | ENST00000498330 | 559 | No protein | - |
| STAT3-233 | ENST00000678108 | 1927 | No protein | - |
| STAT3-207 | ENST00000478276 | 186 | No protein | - |
| STAT3-235 | ENST00000678529 | 2118 | No protein | - |
| STAT3-210 | ENST00000585360 | 517 | No protein | - |
| STAT3-213 | ENST00000590776 | 1483 | No protein | - |
Phenotypes
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| 61951 | Colitis, Ulcerative | 1E-10 | 28067908 |
| 62123 | Crohn Disease | 7E-12 | 18587394 |
| 73876 | Crohn Disease | 2E-15 | 23266558 |
| 76471 | Inflammatory Bowel Diseases | 2E-17 | 28067908 |
| 77738 | Inflammatory Bowel Diseases | 2E-9 | 27569725 |
| 79266 | Inflammatory Bowel Diseases | 1E-22 | 26192919 |
| 84002 | Colitis, Ulcerative | 7E-11 | 26192919 |
| 85343 | Multiple Sclerosis | 3E-10 | 20159113 |
| 85670 | Dermatitis, Atopic | 8E-6 | 25865352 |
| 86826 | Dermatitis, Atopic | 3E-6 | 26482879 |
| 89412 | Inflammatory Bowel Diseases | 6E-22 | 23128233 |
| 97123 | Multiple Sclerosis | 4E-8 | 22190364 |
| 100443 | Multiple Sclerosis | 2E-10 | 21833088 |
| 103069 | Dermatitis, Atopic | 1E-7 | 26482879 |
| 103375 | Crohn Disease | 1E-16 | 26192919 |
| 105134 | Crohn Disease | 2E-11 | 28067908 |
| 107132 | Crohn Disease | 1E-14 | 26192919 |
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
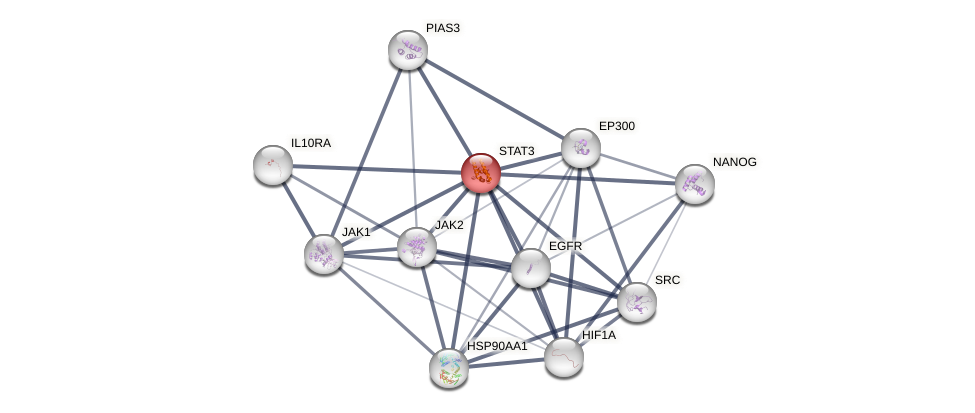
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000168610 | homo_sapiens | ENSP00000300134 | 36.954 | 21.4876 | homo_sapiens | ENSP00000264657 | 40.6493 | 23.6364 |
| ENSG00000168610 | homo_sapiens | ENSP00000293328 | 44.5997 | 28.0813 | homo_sapiens | ENSP00000264657 | 45.5844 | 28.7013 |
| ENSG00000168610 | homo_sapiens | ENSP00000468749 | 44.0806 | 27.9597 | homo_sapiens | ENSP00000264657 | 45.4545 | 28.8312 |
| ENSG00000168610 | homo_sapiens | ENSP00000315768 | 51.1163 | 34.0776 | homo_sapiens | ENSP00000264657 | 56.4935 | 37.6623 |
| ENSG00000168610 | homo_sapiens | ENSP00000376134 | 67.6471 | 48.1283 | homo_sapiens | ENSP00000264657 | 65.7143 | 46.7532 |
| ENSG00000168610 | homo_sapiens | ENSP00000354394 | 72 | 52.1333 | homo_sapiens | ENSP00000264657 | 70.1299 | 50.7792 |
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 100 | 100 | nomascus_leucogenys | ENSNLEP00000014018 | 100 | 100 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 96.6234 | 95.974 | pan_paniscus | ENSPPAP00000035951 | 95.2625 | 94.6223 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 100 | 100 | pongo_abelii | ENSPPYP00000009436 | 100 | 100 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 96.6234 | 95.974 | pan_troglodytes | ENSPTRP00000091410 | 94.1772 | 93.5443 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 96.6234 | 95.974 | gorilla_gorilla | ENSGGOP00000020538 | 95.2625 | 94.6223 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 98.8312 | 98.1818 | ornithorhynchus_anatinus | ENSOANP00000051766 | 98.8312 | 98.1818 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 99.7403 | 99.2208 | sarcophilus_harrisii | ENSSHAP00000017457 | 99.6109 | 99.0921 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 84.8052 | 84.2857 | notamacropus_eugenii | ENSMEUP00000006101 | 84.9155 | 84.3953 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 67.013 | 66.8831 | choloepus_hoffmanni | ENSCHOP00000008280 | 67.1001 | 66.9701 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 100 | 99.8701 | dasypus_novemcinctus | ENSDNOP00000001136 | 100 | 99.8701 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 62.987 | 62.987 | erinaceus_europaeus | ENSEEUP00000003488 | 62.7426 | 62.7426 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 70.2597 | 69.8701 | echinops_telfairi | ENSETEP00000006941 | 85.6013 | 85.1266 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 98.5714 | 98.3117 | callithrix_jacchus | ENSCJAP00000000719 | 98.4436 | 98.1842 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 97.2727 | 96.2338 | cercocebus_atys | ENSCATP00000017410 | 96.3964 | 95.3668 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 100 | 100 | macaca_fascicularis | ENSMFAP00000002773 | 100 | 100 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 100 | 100 | macaca_mulatta | ENSMMUP00000043986 | 88.4041 | 88.4041 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 85.7143 | 85.7143 | macaca_nemestrina | ENSMNEP00000019009 | 100 | 100 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 100 | 100 | papio_anubis | ENSPANP00000020703 | 100 | 100 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 90 | 90 | mandrillus_leucophaeus | ENSMLEP00000012622 | 100 | 100 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 97.6623 | 97.6623 | tursiops_truncatus | ENSTTRP00000006780 | 98.0443 | 98.0443 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 99.8701 | 99.4805 | vulpes_vulpes | ENSVVUP00000032798 | 97.2187 | 96.8394 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 100 | 99.8701 | mus_spretus | MGP_SPRETEiJ_P0031357 | 100 | 99.8701 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 100 | 100 | equus_asinus | ENSEASP00005014293 | 100 | 100 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 99.8701 | 99.8701 | capra_hircus | ENSCHIP00000022282 | 99.8701 | 99.8701 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 99.6104 | 99.3506 | loxodonta_africana | ENSLAFP00000025834 | 99.3523 | 99.0933 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 100 | 100 | panthera_leo | ENSPLOP00000009624 | 100 | 100 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 100 | 100 | ailuropoda_melanoleuca | ENSAMEP00000006689 | 87.8995 | 87.8995 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 99.8701 | 99.7403 | bos_indicus_hybrid | ENSBIXP00005045480 | 99.8701 | 99.7403 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 100 | 99.8701 | oryctolagus_cuniculus | ENSOCUP00000009494 | 100 | 99.8701 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.8571 | 92.8571 | equus_caballus | ENSECAP00000076422 | 100 | 100 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 99.8701 | 99.4805 | canis_lupus_familiaris | ENSCAFP00845012818 | 99.8701 | 99.4805 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 100 | 100 | panthera_pardus | ENSPPRP00000020146 | 100 | 100 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 100 | 100 | marmota_marmota_marmota | ENSMMMP00000006559 | 100 | 100 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 100 | 100 | balaenoptera_musculus | ENSBMSP00010027007 | 88.4041 | 88.4041 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 99.7403 | 99.3506 | cavia_porcellus | ENSCPOP00000025514 | 99.7403 | 99.3506 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 100 | 100 | felis_catus | ENSFCAP00000001991 | 100 | 100 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 100 | 100 | ursus_americanus | ENSUAMP00000008772 | 100 | 100 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 99.7403 | 99.2208 | monodelphis_domestica | ENSMODP00000052576 | 95.4037 | 94.9068 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 98.1818 | 98.1818 | bison_bison_bison | ENSBBBP00000018228 | 99.8679 | 99.8679 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 100 | 99.8701 | monodon_monoceros | ENSMMNP00015025215 | 100 | 99.8701 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 97.2727 | 96.7533 | sus_scrofa | ENSSSCP00000018439 | 97.0207 | 96.5026 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 100 | 99.6104 | microcebus_murinus | ENSMICP00000039856 | 100 | 99.6104 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 80.1299 | 74.9351 | octodon_degus | ENSODEP00000008783 | 87.2702 | 81.6124 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 99.7403 | 99.0909 | octodon_degus | ENSODEP00000024549 | 99.7403 | 99.0909 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 98.7013 | 98.7013 | jaculus_jaculus | ENSJJAP00000024001 | 100 | 100 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 98.1818 | 98.1818 | ovis_aries_rambouillet | ENSOARP00020031000 | 99.8679 | 99.8679 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 100 | 100 | ursus_maritimus | ENSUMAP00000034771 | 91.8854 | 91.8854 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 86.4935 | 86.1039 | ochotona_princeps | ENSOPRP00000007666 | 86.4935 | 86.1039 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 100 | 99.8701 | delphinapterus_leucas | ENSDLEP00000012125 | 100 | 99.8701 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 100 | 100 | physeter_catodon | ENSPCTP00005013731 | 96.1298 | 96.1298 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 100 | 99.8701 | mus_caroli | MGP_CAROLIEiJ_P0029936 | 100 | 99.8701 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 100 | 99.8701 | mus_musculus | ENSMUSP00000120152 | 100 | 99.8701 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 99.7403 | 99.6104 | dipodomys_ordii | ENSDORP00000024461 | 99.7403 | 99.6104 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 99.8701 | 99.7403 | mustela_putorius_furo | ENSMPUP00000010432 | 99.8701 | 99.7403 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 100 | 100 | aotus_nancymaae | ENSANAP00000037142 | 95.6522 | 95.6522 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 93.6364 | 93.6364 | saimiri_boliviensis_boliviensis | ENSSBOP00000037847 | 100 | 100 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 100 | 100 | chlorocebus_sabaeus | ENSCSAP00000014828 | 100 | 100 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 55.0649 | 54.8052 | tupaia_belangeri | ENSTBEP00000004768 | 54.9223 | 54.6632 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 95.8442 | 95.0649 | mesocricetus_auratus | ENSMAUP00000007036 | 97.3615 | 96.5699 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 100 | 99.8701 | phocoena_sinus | ENSPSNP00000022904 | 100 | 99.8701 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 100 | 100 | camelus_dromedarius | ENSCDRP00005007495 | 100 | 100 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 55.974 | 55.3247 | carlito_syrichta | ENSTSYP00000011433 | 97.0721 | 95.9459 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 100 | 100 | panthera_tigris_altaica | ENSPTIP00000007211 | 100 | 100 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 100 | 99.7403 | rattus_norvegicus | ENSRNOP00000092154 | 98.0892 | 97.8344 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 98.0519 | 97.4026 | prolemur_simus | ENSPSMP00000005465 | 97.9248 | 97.2763 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 98.4416 | 98.3117 | microtus_ochrogaster | ENSMOCP00000010986 | 99.2147 | 99.0838 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 99.7403 | 99.6104 | sorex_araneus | ENSSARP00000012881 | 99.4819 | 99.3523 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 100 | 99.8701 | peromyscus_maniculatus_bairdii | ENSPEMP00000016743 | 100 | 99.8701 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 99.7403 | 99.4805 | bos_mutus | ENSBMUP00000021804 | 99.7403 | 99.4805 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 100 | 99.8701 | mus_spicilegus | ENSMSIP00000005871 | 100 | 99.8701 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 86.8831 | 85.5844 | cricetulus_griseus_chok1gshd | ENSCGRP00001011560 | 98.6726 | 97.1976 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 97.2727 | 97.2727 | pteropus_vampyrus | ENSPVAP00000003681 | 97.2727 | 97.2727 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 100 | 100 | myotis_lucifugus | ENSMLUP00000015910 | 99.6119 | 99.6119 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 99.8701 | 99.3506 | vombatus_ursinus | ENSVURP00010028536 | 99.7406 | 99.2218 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 100 | 100 | rhinolophus_ferrumequinum | ENSRFEP00010011810 | 100 | 100 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 97.2727 | 97.1429 | neovison_vison | ENSNVIP00000016057 | 99.8667 | 99.7333 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 99.8701 | 99.7403 | bos_taurus | ENSBTAP00000028687 | 99.8701 | 99.7403 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 99.6104 | 99.0909 | heterocephalus_glaber_female | ENSHGLP00000015256 | 88.3641 | 87.9032 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 90.7792 | 89.2208 | rhinopithecus_bieti | ENSRBIP00000008356 | 90.7792 | 89.2208 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 99.8701 | 99.4805 | canis_lupus_dingo | ENSCAFP00020016836 | 99.8701 | 99.4805 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 100 | 100 | sciurus_vulgaris | ENSSVLP00005005543 | 100 | 100 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 99.8701 | 99.2208 | phascolarctos_cinereus | ENSPCIP00000034506 | 99.7406 | 99.0921 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 63.6364 | 63.5065 | procavia_capensis | ENSPCAP00000009640 | 67.3077 | 67.1703 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 100 | 100 | nannospalax_galili | ENSNGAP00000002603 | 100 | 100 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 99.8701 | 99.7403 | bos_grunniens | ENSBGRP00000016397 | 99.8701 | 99.7403 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 99.8701 | 99.3506 | chinchilla_lanigera | ENSCLAP00000021498 | 97.7128 | 97.2046 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 99.8701 | 99.8701 | cervus_hanglu_yarkandensis | ENSCHYP00000014139 | 99.8701 | 99.8701 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 99.8701 | 99.8701 | catagonus_wagneri | ENSCWAP00000018926 | 99.8701 | 99.8701 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 91.1688 | 89.8701 | ictidomys_tridecemlineatus | ENSSTOP00000006050 | 93.8503 | 92.5134 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 99.8701 | 99.6104 | otolemur_garnettii | ENSOGAP00000001349 | 99.8701 | 99.6104 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 100 | 100 | cebus_imitator | ENSCCAP00000017914 | 100 | 100 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 100 | 99.8701 | urocitellus_parryii | ENSUPAP00010011945 | 100 | 99.8701 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 99.8701 | 99.8701 | moschus_moschiferus | ENSMMSP00000021197 | 99.8701 | 99.8701 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 98.4416 | 97.9221 | rhinopithecus_roxellana | ENSRROP00000018412 | 94.5137 | 94.015 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 100 | 99.7403 | mus_pahari | MGP_PahariEiJ_P0036659 | 100 | 99.7403 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 93.7662 | 91.9481 | propithecus_coquereli | ENSPCOP00000030268 | 90.7035 | 88.9447 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 70 | 54.5455 | petromyzon_marinus | ENSPMAP00000000297 | 70.6422 | 55.0459 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 12.8571 | 10.2597 | eptatretus_burgeri | ENSEBUP00000004966 | 56.5714 | 45.1429 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.4675 | 86.4935 | callorhinchus_milii | ENSCMIP00000006620 | 92.4675 | 86.4935 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 88.5714 | 84.9351 | latimeria_chalumnae | ENSLACP00000013656 | 89.384 | 85.7143 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 93.7662 | 89.6104 | lepisosteus_oculatus | ENSLOCP00000015422 | 91.5082 | 87.4525 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 91.6883 | 86.3636 | clupea_harengus | ENSCHAP00000008527 | 88.3605 | 83.229 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.987 | 88.3117 | danio_rerio | ENSDARP00000095292 | 88.8337 | 84.3672 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 93.1169 | 88.0519 | carassius_auratus | ENSCARP00000037575 | 88.4094 | 83.6005 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.8571 | 88.1818 | carassius_auratus | ENSCARP00000016366 | 88.7097 | 84.2432 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.5974 | 88.3117 | astyanax_mexicanus | ENSAMXP00000050869 | 90.4822 | 86.2944 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.2078 | 87.2727 | ictalurus_punctatus | ENSIPUP00000030311 | 90.1015 | 85.2792 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 91.2987 | 86.2338 | esox_lucius | ENSELUP00000019487 | 80.8046 | 76.3218 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 72.3377 | 66.6234 | electrophorus_electricus | ENSEEEP00000008570 | 82.5185 | 76 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.987 | 87.5325 | oncorhynchus_kisutch | ENSOKIP00005085895 | 87.8528 | 82.6994 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.8571 | 87.4026 | oncorhynchus_kisutch | ENSOKIP00005106984 | 89.4869 | 84.2303 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.8571 | 87.4026 | oncorhynchus_mykiss | ENSOMYP00000013242 | 89.4869 | 84.2303 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.8571 | 87.2727 | oncorhynchus_mykiss | ENSOMYP00000022925 | 88.3807 | 83.0655 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.8571 | 87.1429 | salmo_salar | ENSSSAP00000007429 | 87.4083 | 82.0293 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 89.7403 | 83.8961 | salmo_salar | ENSSSAP00000021394 | 85.5198 | 79.9505 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 91.6883 | 85.974 | salmo_trutta | ENSSTUP00000047947 | 87.5931 | 82.134 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.8571 | 87.1429 | salmo_trutta | ENSSTUP00000090139 | 88.3807 | 82.9419 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 93.2467 | 87.7922 | gadus_morhua | ENSGMOP00000068902 | 90.201 | 84.9246 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.0779 | 86.6234 | fundulus_heteroclitus | ENSFHEP00000028012 | 90.0889 | 84.7522 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 89.8701 | 84.4156 | poecilia_reticulata | ENSPREP00000014658 | 83.8788 | 78.7879 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.2078 | 87.2727 | xiphophorus_maculatus | ENSXMAP00000009027 | 90.216 | 85.3876 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.4675 | 87.7922 | oryzias_latipes | ENSORLP00000005087 | 90.7006 | 86.1146 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.3377 | 87.4026 | cyclopterus_lumpus | ENSCLMP00005031398 | 90.6888 | 85.8418 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 91.4286 | 86.2338 | oreochromis_niloticus | ENSONIP00000039168 | 88.5535 | 83.522 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 91.6883 | 87.013 | haplochromis_burtoni | ENSHBUP00000000954 | 85.3688 | 81.0157 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.4675 | 87.5325 | astatotilapia_calliptera | ENSACLP00000004595 | 88.8889 | 84.1448 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.5974 | 87.7922 | sparus_aurata | ENSSAUP00010053822 | 90.9439 | 86.2245 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.4675 | 87.5325 | lates_calcarifer | ENSLCAP00010015319 | 89.335 | 84.5671 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 97.9221 | 96.1039 | xenopus_tropicalis | ENSXETP00000103484 | 98.0494 | 96.2289 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 98.8312 | 98.1818 | chrysemys_picta_bellii | ENSCPBP00000023238 | 98.703 | 98.0545 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 98.3117 | 97.6623 | sphenodon_punctatus | ENSSPUP00000007764 | 86.9116 | 86.3375 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 99.0909 | 98.4416 | crocodylus_porosus | ENSCPRP00005008425 | 98.9624 | 98.3139 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 60.9091 | 59.4805 | laticauda_laticaudata | ENSLLTP00000016227 | 98.5294 | 96.2185 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 28.0519 | 26.7532 | laticauda_laticaudata | ENSLLTP00000005065 | 96.4286 | 91.9643 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 96.4935 | 94.5455 | notechis_scutatus | ENSNSUP00000012672 | 96.2435 | 94.3005 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 98.961 | 97.2727 | pseudonaja_textilis | ENSPTXP00000005304 | 98.7047 | 97.0207 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 98.5714 | 97.6623 | anas_platyrhynchos_platyrhynchos | ENSAPLP00000007496 | 80.5732 | 79.8301 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 98.1818 | 97.4026 | gallus_gallus | ENSGALP00010039261 | 98.5658 | 97.7836 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 98.5714 | 97.6623 | meleagris_gallopavo | ENSMGAP00000003284 | 98.4436 | 97.5357 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 98.5714 | 97.9221 | serinus_canaria | ENSSCAP00000009103 | 98.4436 | 97.7951 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 98.5714 | 97.9221 | parus_major | ENSPMJP00000001401 | 98.4436 | 97.7951 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 98.4416 | 96.4935 | anolis_carolinensis | ENSACAP00000017673 | 98.0595 | 96.119 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.4675 | 87.5325 | neolamprologus_brichardi | ENSNBRP00000022460 | 93.1937 | 88.2199 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 98.961 | 97.1429 | naja_naja | ENSNNAP00000009539 | 98.7047 | 96.8912 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 98.0519 | 96.2338 | podarcis_muralis | ENSPMRP00000025003 | 89.3491 | 87.6923 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 98.5714 | 97.7922 | geospiza_fortis | ENSGFOP00000006579 | 98.4436 | 97.6654 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 97.013 | 96.3636 | taeniopygia_guttata | ENSTGUP00000002670 | 92.1085 | 91.492 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 87.9221 | 84.6753 | erpetoichthys_calabaricus | ENSECRP00000017009 | 95.218 | 91.7018 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.3377 | 86.7533 | kryptolebias_marmoratus | ENSKMAP00000026789 | 90.3431 | 84.8793 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 98.7013 | 97.1429 | salvator_merianae | ENSSMRP00000002976 | 88.993 | 87.5878 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.2078 | 87.2727 | oryzias_javanicus | ENSOJAP00000030843 | 78.714 | 74.5011 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 87.9221 | 83.2467 | amphilophus_citrinellus | ENSACIP00000014505 | 92.8669 | 87.9287 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 91.9481 | 86.8831 | dicentrarchus_labrax | ENSDLAP00005047080 | 87.8412 | 83.0025 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.0779 | 87.1429 | amphiprion_percula | ENSAPEP00000025897 | 89.7468 | 84.9367 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.4675 | 87.7922 | oryzias_sinensis | ENSOSIP00000001901 | 90.7006 | 86.1146 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 86.4935 | 85.3247 | pelodiscus_sinensis | ENSPSIP00000018063 | 94.2009 | 92.9279 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 98.8312 | 98.4416 | gopherus_evgoodei | ENSGEVP00005011037 | 93.8348 | 93.4649 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 98.8312 | 98.4416 | chelonoidis_abingdonii | ENSCABP00000001766 | 98.703 | 98.3139 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.4675 | 87.7922 | seriola_dumerili | ENSSDUP00000023515 | 90.9323 | 86.3346 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.3377 | 87.2727 | cottoperca_gobio | ENSCGOP00000029011 | 90.6888 | 85.7143 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 98.7013 | 98.0519 | struthio_camelus_australis | ENSSCUP00000016682 | 98.5733 | 97.9248 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 98.5714 | 97.6623 | anser_brachyrhynchus | ENSABRP00000014287 | 90.0356 | 89.2052 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.7273 | 87.5325 | gasterosteus_aculeatus | ENSGACP00000011388 | 88.5856 | 83.6228 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.3377 | 87.6623 | cynoglossus_semilaevis | ENSCSEP00000026841 | 90.8046 | 86.2069 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.0779 | 86.7533 | poecilia_formosa | ENSPFOP00000013256 | 90.0889 | 84.8793 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 88.8312 | 84.2857 | poecilia_formosa | ENSPFOP00000012972 | 93.8272 | 89.0261 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 93.1169 | 88.0519 | myripristis_murdjan | ENSMMDP00005008610 | 90.9898 | 86.0406 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.5974 | 87.6623 | sander_lucioperca | ENSSLUP00000022006 | 90.9439 | 86.0969 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 98.3117 | 97.5325 | strigops_habroptila | ENSSHBP00005014475 | 98.1842 | 97.406 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.5974 | 87.9221 | amphiprion_ocellaris | ENSAOCP00000010154 | 90.9439 | 86.352 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 97.7922 | 96.8831 | terrapene_carolina_triunguis | ENSTMTP00000007663 | 92.3926 | 91.5337 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.987 | 87.4026 | hucho_hucho | ENSHHUP00000086381 | 89.612 | 84.2303 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 91.5584 | 85.5844 | hucho_hucho | ENSHHUP00000043592 | 87.6866 | 81.9652 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.0779 | 87.4026 | betta_splendens | ENSBSLP00000008669 | 90.0889 | 85.5146 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.5974 | 88.1818 | pygocentrus_nattereri | ENSPNAP00000028519 | 90.1391 | 85.8407 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.3377 | 87.9221 | anabas_testudineus | ENSATEP00000003147 | 90.6888 | 86.352 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.4675 | 87.7922 | seriola_lalandi_dorsalis | ENSSLDP00000010111 | 90.9323 | 86.3346 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 15.1948 | 14.1558 | seriola_lalandi_dorsalis | ENSSLDP00000004146 | 90 | 83.8462 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 87.1429 | 82.4675 | labrus_bergylta | ENSLBEP00000005025 | 94.774 | 89.6893 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.4675 | 87.5325 | pundamilia_nyererei | ENSPNYP00000013667 | 90.8163 | 85.9694 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.4675 | 88.0519 | mastacembelus_armatus | ENSMAMP00000049403 | 90.8163 | 86.4796 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.5974 | 87.6623 | stegastes_partitus | ENSSPAP00000027206 | 90.9439 | 86.0969 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.8571 | 87.1429 | oncorhynchus_tshawytscha | ENSOTSP00005100638 | 89.2634 | 83.7703 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 88.1818 | 83.1169 | oncorhynchus_tshawytscha | ENSOTSP00005056882 | 87.7261 | 82.6873 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.5974 | 87.9221 | acanthochromis_polyacanthus | ENSAPOP00000002542 | 90.828 | 86.242 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 98.5714 | 97.7922 | aquila_chrysaetos_chrysaetos | ENSACCP00020002574 | 98.4436 | 97.6654 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.3377 | 87.2727 | nothobranchius_furzeri | ENSNFUP00015004982 | 87.0257 | 82.2521 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.0779 | 86.8831 | cyprinodon_variegatus | ENSCVAP00000019613 | 88.625 | 83.625 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.4675 | 87.1429 | denticeps_clupeoides | ENSDCDP00000006544 | 90.5852 | 85.369 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.5974 | 87.6623 | larimichthys_crocea | ENSLCRP00005002838 | 87.3774 | 82.7206 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.3377 | 87.1429 | takifugu_rubripes | ENSTRUP00000056136 | 90.6888 | 85.5867 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 98.3117 | 97.5325 | ficedula_albicollis | ENSFALP00000019857 | 96.066 | 95.3046 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 91.8182 | 87.1429 | hippocampus_comes | ENSHCOP00000020969 | 89.1551 | 84.6154 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.4675 | 87.7922 | cyprinus_carpio_carpio | ENSCCRP00000080760 | 89.7856 | 85.2459 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.987 | 88.1818 | cyprinus_carpio_carpio | ENSCCRP00000017507 | 88.8337 | 84.2432 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 98.4416 | 97.6623 | coturnix_japonica | ENSCJPP00005005041 | 98.3139 | 97.5357 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.0779 | 86.7533 | poecilia_latipinna | ENSPLAP00000013223 | 90.0889 | 84.8793 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.8571 | 87.7922 | sinocyclocheilus_grahami | ENSSGRP00000085654 | 88.1628 | 83.3539 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 88.7013 | 83.7662 | sinocyclocheilus_grahami | ENSSGRP00000025008 | 88.357 | 83.4411 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.4675 | 87.5325 | maylandia_zebra | ENSMZEP00005030472 | 90.8163 | 85.9694 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 35.3247 | 33.7662 | tetraodon_nigroviridis | ENSTNIP00000014657 | 84.7352 | 80.9969 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 34.026 | 31.2987 | tetraodon_nigroviridis | ENSTNIP00000022932 | 90.3448 | 83.1034 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.7273 | 88.5714 | paramormyrops_kingsleyae | ENSPKIP00000013657 | 90.9554 | 86.879 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 90.2597 | 84.6753 | paramormyrops_kingsleyae | ENSPKIP00000001757 | 87.202 | 81.8068 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 90.5195 | 84.8052 | scleropages_formosus | ENSSFOP00015045289 | 84.5874 | 79.2476 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.8571 | 88.3117 | scleropages_formosus | ENSSFOP00015015162 | 88.4901 | 84.1584 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 97.1429 | 94.4156 | leptobrachium_leishanense | ENSLLEP00000044040 | 97.3958 | 94.6615 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 92.7273 | 87.7922 | scophthalmus_maximus | ENSSMAP00000011378 | 91.0714 | 86.2245 |
| ENSG00000168610 | homo_sapiens | ENSP00000264657 | 90.6494 | 85.8442 | oryzias_melastigma | ENSOMEP00000021350 | 86.6005 | 82.0099 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0000122 | involved_in negative regulation of transcription by RNA polymerase II | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0000785 | part_of chromatin | 2 | IDA,ISA | Homo_sapiens(9606) | Component |
| GO:0000976 | enables transcription cis-regulatory region binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0000978 | enables RNA polymerase II cis-regulatory region sequence-specific DNA binding | 2 | IBA,IDA | Homo_sapiens(9606) | Function |
| GO:0000981 | enables DNA-binding transcription factor activity, RNA polymerase II-specific | 3 | IBA,IDA,ISA | Homo_sapiens(9606) | Function |
| GO:0001228 | enables DNA-binding transcription activator activity, RNA polymerase II-specific | 3 | IDA,IGI,IMP | Homo_sapiens(9606) | Function |
| GO:0001659 | involved_in temperature homeostasis | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0001754 | involved_in eye photoreceptor cell differentiation | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0003677 | enables DNA binding | 1 | ISS | Homo_sapiens(9606) | Function |
| GO:0003700 | enables DNA-binding transcription factor activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0004879 | enables nuclear receptor activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0005102 | enables signaling receptor binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005634 | is_active_in nucleus | 5 | IC,IDA,ISS | Homo_sapiens(9606) | Component |
| GO:0005654 | located_in nucleoplasm | 2 | IDA,TAS | Homo_sapiens(9606) | Component |
| GO:0005667 | part_of transcription regulator complex | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005737 | located_in cytoplasm | 2 | IDA,ISS | Homo_sapiens(9606) | Component |
| GO:0005829 | located_in cytosol | 2 | IDA,TAS | Homo_sapiens(9606) | Component |
| GO:0005886 | located_in plasma membrane | 1 | ISS | Homo_sapiens(9606) | Component |
| GO:0006355 | involved_in regulation of DNA-templated transcription | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0006357 | involved_in regulation of transcription by RNA polymerase II | 2 | IBA,ISS | Homo_sapiens(9606) | Process |
| GO:0006606 | involved_in protein import into nucleus | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0006952 | involved_in defense response | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0006954 | involved_in inflammatory response | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0007165 | involved_in signal transduction | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0007179 | involved_in transforming growth factor beta receptor signaling pathway | 1 | IGI | Homo_sapiens(9606) | Process |
| GO:0007259 | involved_in receptor signaling pathway via JAK-STAT | 4 | IBA,IDA,NAS,TAS | Homo_sapiens(9606) | Process |
| GO:0007399 | involved_in nervous system development | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0008283 | involved_in cell population proliferation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0008285 | involved_in negative regulation of cell population proliferation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0010467 | involved_in gene expression | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0010507 | involved_in negative regulation of autophagy | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0010628 | involved_in positive regulation of gene expression | 2 | IDA,IMP | Homo_sapiens(9606) | Process |
| GO:0010629 | involved_in negative regulation of gene expression | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0014065 | involved_in phosphatidylinositol 3-kinase signaling | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0016310 | involved_in phosphorylation | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0019221 | involved_in cytokine-mediated signaling pathway | 1 | NAS | Homo_sapiens(9606) | Process |
| GO:0019901 | enables protein kinase binding | 1 | ISS | Homo_sapiens(9606) | Function |
| GO:0019903 | enables protein phosphatase binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0019953 | involved_in sexual reproduction | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0030154 | involved_in cell differentiation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0030335 | involved_in positive regulation of cell migration | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0030522 | involved_in intracellular receptor signaling pathway | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0031490 | enables chromatin DNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0032355 | involved_in response to estradiol | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0032731 | involved_in positive regulation of interleukin-1 beta production | 1 | IGI | Homo_sapiens(9606) | Process |
| GO:0032733 | involved_in positive regulation of interleukin-10 production | 1 | IGI | Homo_sapiens(9606) | Process |
| GO:0032755 | involved_in positive regulation of interleukin-6 production | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0032757 | involved_in positive regulation of interleukin-8 production | 1 | IGI | Homo_sapiens(9606) | Process |
| GO:0032760 | involved_in positive regulation of tumor necrosis factor production | 1 | IGI | Homo_sapiens(9606) | Process |
| GO:0032870 | involved_in cellular response to hormone stimulus | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0033210 | involved_in leptin-mediated signaling pathway | 2 | IBA,IDA | Homo_sapiens(9606) | Process |
| GO:0035019 | involved_in somatic stem cell population maintenance | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0035591 | enables signaling adaptor activity | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0035723 | involved_in interleukin-15-mediated signaling pathway | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0038110 | involved_in interleukin-2-mediated signaling pathway | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0038113 | involved_in interleukin-9-mediated signaling pathway | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0038154 | involved_in interleukin-11-mediated signaling pathway | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0040014 | involved_in regulation of multicellular organism growth | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0042127 | involved_in regulation of cell population proliferation | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0042593 | involved_in glucose homeostasis | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0042755 | involved_in eating behavior | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0042789 | involved_in mRNA transcription by RNA polymerase II | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0042802 | enables identical protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0042803 | enables protein homodimerization activity | 1 | ISS | Homo_sapiens(9606) | Function |
| GO:0043434 | involved_in response to peptide hormone | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0044320 | involved_in cellular response to leptin stimulus | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0044321 | involved_in response to leptin | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0045648 | involved_in positive regulation of erythrocyte differentiation | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0045747 | involved_in positive regulation of Notch signaling pathway | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0045766 | involved_in positive regulation of angiogenesis | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0045820 | involved_in negative regulation of glycolytic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0045893 | involved_in positive regulation of DNA-templated transcription | 2 | IDA,ISS | Homo_sapiens(9606) | Process |
| GO:0045944 | involved_in positive regulation of transcription by RNA polymerase II | 4 | IDA,IGI,IMP,NAS | Homo_sapiens(9606) | Process |
| GO:0046983 | enables protein dimerization activity | 1 | ISS | Homo_sapiens(9606) | Function |
| GO:0048708 | involved_in astrocyte differentiation | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0050728 | involved_in negative regulation of inflammatory response | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0051092 | involved_in positive regulation of NF-kappaB transcription factor activity | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0051726 | involved_in regulation of cell cycle | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0060019 | involved_in radial glial cell differentiation | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0060221 | involved_in retinal rod cell differentiation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0060259 | involved_in regulation of feeding behavior | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0060396 | involved_in growth hormone receptor signaling pathway | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0060397 | involved_in growth hormone receptor signaling pathway via JAK-STAT | 3 | IBA,IDA,ISS | Homo_sapiens(9606) | Process |
| GO:0061629 | enables RNA polymerase II-specific DNA-binding transcription factor binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0070102 | involved_in interleukin-6-mediated signaling pathway | 2 | IDA,IMP | Homo_sapiens(9606) | Process |
| GO:0070878 | enables primary miRNA binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0072538 | involved_in T-helper 17 type immune response | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0072540 | involved_in T-helper 17 cell lineage commitment | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0090575 | part_of RNA polymerase II transcription regulator complex | 4 | IC,IMP,IPI,NAS | Homo_sapiens(9606) | Component |
| GO:0097009 | involved_in energy homeostasis | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0097398 | involved_in cellular response to interleukin-17 | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0106015 | involved_in negative regulation of inflammatory response to wounding | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0140297 | enables DNA-binding transcription factor binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0140610 | enables RNA sequestering activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:1900017 | involved_in positive regulation of cytokine production involved in inflammatory response | 1 | IGI | Homo_sapiens(9606) | Process |
| GO:1902895 | involved_in positive regulation of miRNA transcription | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:1904685 | involved_in positive regulation of metalloendopeptidase activity | 1 | IGI | Homo_sapiens(9606) | Process |
| GO:1905564 | involved_in positive regulation of vascular endothelial cell proliferation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:2000635 | involved_in negative regulation of primary miRNA processing | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:2000737 | involved_in negative regulation of stem cell differentiation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:2001223 | involved_in negative regulation of neuron migration | 1 | IEA | Homo_sapiens(9606) | Process |