| RBP Type |
Canonical_RBPs |
| Diseases |
N.A. |
| Drug |
N.A. |
| Main interacting RNAs |
N.A. |
| Moonlighting functions |
E3 ligase |
| Localizations |
N.A. |
|
BulkPerturb-seq
|
|
| Ensembl ID | ENSG00000167333 | Gene ID |
55128 |
Accession |
21161 |
| Symbol |
TRIM68 |
Alias |
SS56;GC109;SS-56;RNF137 |
Full Name |
tripartite motif containing 68 |
| Status |
Confidence |
Length |
9560 bases |
Strand |
Minus strand |
| Position |
11 : 4598672 - 4608231 |
RNA binding domain |
SPRY |
| Summary |
|
RNA binding domains (RBDs)

| Protein |
Domain |
Pfam ID |
E-value |
Domain number |
| ENSP00000300747 |
SPRY |
PF00622 |
5.8e-14 |
1 |
RNA binding proteomes (RBPomes)
| Pubmed ID |
Full Name |
Cell |
Author |
Time |
Doi |
Literatures on RNA binding capacity
| Pubmed ID |
Title |
Author |
Time |
Journal |
| Name |
Transcript ID |
bp |
Protein |
Translation ID |
| TRIM68-201 |
ENST00000300747 |
3324 |
485aa |
ENSP00000300747 |
| TRIM68-203 |
ENST00000531101 |
3187 |
187aa |
ENSP00000431783 |
| TRIM68-202 |
ENST00000526337 |
874 |
142aa |
ENSP00000434681 |
| TRIM68-205 |
ENST00000531717 |
566 |
No protein |
- |
| TRIM68-204 |
ENST00000531644 |
553 |
No protein |
- |
| TRIM68-206 |
ENST00000532108 |
1462 |
No protein |
- |
| TRIM68-207 |
ENST00000533021 |
764 |
198aa |
ENSP00000436112 |
| Pathway ID |
Pathway Name |
Source |
| | |
| ensgID |
Trait |
pValue |
Pubmed ID |
| 18251 | Fetal Hemoglobin | 3.825e-005 | - |
| 18252 | Fetal Hemoglobin | 3.825e-005 | - |
| 18635 | Erythrocyte Indices | 4.933e-009 | - |
| 18636 | Erythrocyte Indices | 4.933e-009 | - |
| 18757 | Erythrocyte Indices | 2.002e-007 | - |
| 18758 | Erythrocyte Indices | 2.002e-007 | - |
| 18908 | Erythrocyte Count | 1.694e-006 | - |
| 18909 | Erythrocyte Count | 1.694e-006 | - |
| ensgID | SNP | Chromosome | Position |
Trait | PubmedID | Or or BEAT |
EFO ID |
|---|
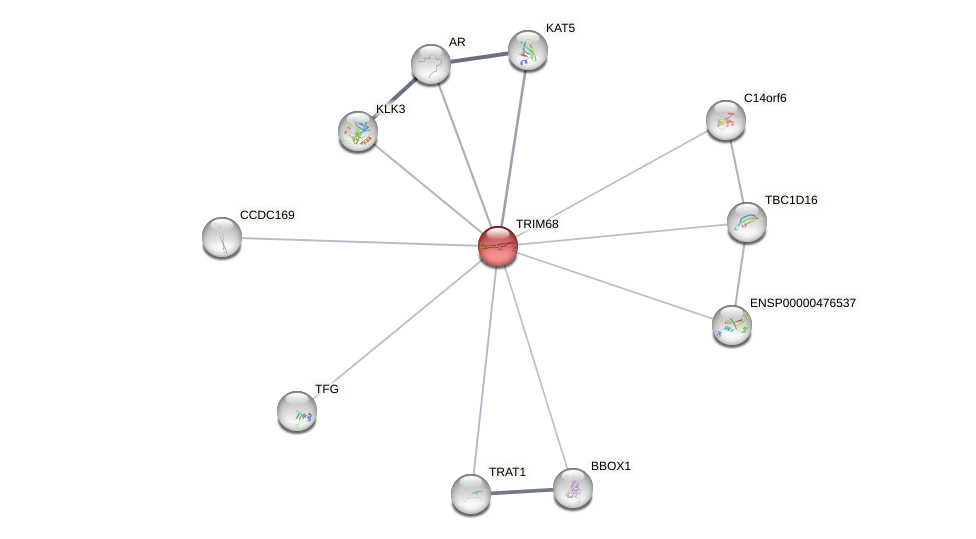
Protein-Protein Interaction (PPI)
| Ensembl ID |
Source |
Target |
| Species |
Protein ID |
Perc_pos |
Perc_id |
Species |
Protein ID |
Perc_pos |
Perc_id |
| Ensembl ID |
Source |
Target |
| Species |
Protein ID |
Perc_pos |
Perc_id |
Species |
Protein ID |
Perc_pos |
Perc_id |
| Go ID |
Go term |
No. evidence |
Entries |
Species |
Category |
| GO:0000209 | involved_in protein polyubiquitination | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0004842 | enables ubiquitin-protein transferase activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005634 | located_in nucleus | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005654 | is_active_in nucleoplasm | 2 | IBA,IDA | Homo_sapiens(9606) | Component |
| GO:0005730 | located_in nucleolus | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005737 | is_active_in cytoplasm | 2 | IBA,IDA | Homo_sapiens(9606) | Component |
| GO:0005794 | located_in Golgi apparatus | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005829 | is_active_in cytosol | 3 | IBA,IDA,TAS | Homo_sapiens(9606) | Component |
| GO:0008270 | enables zinc ion binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0010508 | involved_in positive regulation of autophagy | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0019901 | enables protein kinase binding | 1 | IBA | Homo_sapiens(9606) | Function |
| GO:0032880 | involved_in regulation of protein localization | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0035035 | enables histone acetyltransferase binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0042802 | enables identical protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0042803 | enables protein homodimerization activity | 1 | IBA | Homo_sapiens(9606) | Function |
| GO:0043123 | involved_in positive regulation of I-kappaB kinase/NF-kappaB signaling | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0045087 | involved_in innate immune response | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0046596 | involved_in regulation of viral entry into host cell | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0048471 | located_in perinuclear region of cytoplasm | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0050681 | enables nuclear androgen receptor binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0051092 | involved_in positive regulation of NF-kappaB transcription factor activity | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0051865 | involved_in protein autoubiquitination | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0060765 | involved_in regulation of androgen receptor signaling pathway | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0061630 | enables ubiquitin protein ligase activity | 1 | IBA | Homo_sapiens(9606) | Function |