Welcome to
RBP World!
Functions and Diseases
| RBP Type | Canonical_RBPs |
| Diseases | CHIKVSARS-COV-OC43FlavivirusesHIVRVSARS-COV-2SINVDengue |
| Drug | N.A. |
| Main interacting RNAs | rRNA |
| Moonlighting functions | N.A. |
| Localizations | Stress granucleP-bodyNucleoli rim |
| BulkPerturb-seq | DataSet_01_117 DataSet_01_12 |
Description
| Ensembl ID | ENSG00000165732 | Gene ID | 9188 | Accession | 2744 |
| Symbol | DDX21 | Alias | RH;GUA;GURDB;II/Gu;RHII/Gu;RH-II/GU;gu-alpha;RH-II/GuA | Full Name | DExD-box helicase 21 |
| Status | Confidence | Length | 28934 bases | Strand | Plus strand |
| Position | 10 : 68956135 - 68985068 | RNA binding domain | ResIII , DEAD , GUCT | ||
| Summary | DEAD box proteins, characterized by the conserved motif Asp-Glu-Ala-Asp (DEAD), are putative RNA helicases. They are implicated in a number of cellular processes involving alteration of RNA secondary structure such as translation initiation, nuclear and mitochondrial splicing, and ribosome and spliceosome assembly. Based on their distribution patterns, some members of this family are believed to be involved in embryogenesis, spermatogenesis, and cellular growth and division. This gene encodes a DEAD box protein, which is an antigen recognized by autoimmune antibodies from a patient with watermelon stomach disease. This protein unwinds double-stranded RNA, folds single-stranded RNA, and may play important roles in ribosomal RNA biogenesis, RNA editing, RNA transport, and general transcription. [provided by RefSeq, Jul 2008] | ||||
RNA binding domains (RBDs)
| Protein | Domain | Pfam ID | E-value | Domain number |
|---|---|---|---|---|
| ENSP00000480334 | DEAD | PF00270 | 1.2e-46 | 1 |
| ENSP00000480334 | ResIII | PF04851 | 3.5e-08 | 2 |
| ENSP00000480334 | GUCT | PF08152 | 2.1e-29 | 3 |
| ENSP00000510035 | DEAD | PF00270 | 1.2e-46 | 1 |
| ENSP00000510035 | ResIII | PF04851 | 3.5e-08 | 2 |
| ENSP00000510035 | GUCT | PF08152 | 2.1e-29 | 3 |
| ENSP00000508484 | DEAD | PF00270 | 1.3e-46 | 1 |
| ENSP00000508484 | ResIII | PF04851 | 3.1e-08 | 2 |
| ENSP00000508484 | GUCT | PF08152 | 2.1e-29 | 3 |
| ENSP00000508639 | DEAD | PF00270 | 1.3e-46 | 1 |
| ENSP00000508639 | ResIII | PF04851 | 4e-08 | 2 |
| ENSP00000508639 | GUCT | PF08152 | 6.8e-17 | 3 |
| ENSP00000346120 | DEAD | PF00270 | 1.4e-46 | 1 |
| ENSP00000346120 | ResIII | PF04851 | 4.2e-08 | 2 |
| ENSP00000346120 | GUCT | PF08152 | 2.3e-29 | 3 |
| ENSP00000510328 | DEAD | PF00270 | 1.5e-46 | 1 |
| ENSP00000510328 | ResIII | PF04851 | 3.7e-08 | 2 |
| ENSP00000510328 | GUCT | PF08152 | 2.3e-29 | 3 |
| ENSP00000508611 | DEAD | PF00270 | 1.8e-46 | 1 |
| ENSP00000508611 | ResIII | PF04851 | 4.4e-08 | 2 |
| ENSP00000508611 | GUCT | PF08152 | 2.1e-29 | 3 |
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|---|---|---|---|
| 35336874 | DEAD-Box RNA Helicase 21 (DDX21) Positively Regulates the Replication of Porcine Reproductive and Respiratory Syndrome Virus via Multiple Mechanisms. | Jia Li | 2022-02-24 | Viruses |
| 24721576 | Cellular DDX21 RNA helicase inhibits influenza A virus replication but is counteracted by the viral NS1 protein. | Guifang Chen | 2014-04-09 | Cell host & microbe |
| 31351877 | Activation of PARP-1 by snoRNAs Controls Ribosome Biogenesis and Cell Growth via the RNA Helicase DDX21. | Dae-Seok Kim | 2019-09-19 | Molecular cell |
| 33040459 | EBP2, a novel NPM-ALK-interacting protein in the nucleolus, contributes to the proliferation of ALCL cells by regulating tumor suppressor p53. | Yuki Uchihara | 2021-01-01 | Molecular oncology |
| 28705764 | A Survey of DDX21 Activity During Rev/RRE Complex Formation. | A John Hammond | 2018-02-16 | Journal of molecular biology |
| 31754723 | A double-stranded RNA platform is required for the interaction between a host restriction factor and the NS1 protein of influenza A virus. | Guifang Chen | 2020-01-10 | Nucleic acids research |
| 36608661 | Glucose dissociates DDX21 dimers to regulate mRNA splicing and tissue differentiation. | Weili Miao | 2023-01-05 | Cell |
| 29906500 | Insights into the RNA quadruplex binding specificity of DDX21. | S K Ewan McRae | 2018-09-01 | Biochimica et biophysica acta. General subjects |
| 33505109 | Downregulation of lncRNA HCP5 has inhibitory effects on gastric cancer cells by regulating DDX21 expression. | Kehao Wang | 2021-02-01 | Cytotechnology |
| 35852833 | NAT10 and DDX21 Proteins Interact with RNase H1 and Affect the Performance of Phosphorothioate Oligonucleotides. | Lingdi Zhang | 2022-08-01 | Nucleic acid therapeutics |
| 35328433 | Fragments of rDNA Genes Scattered over the Human Genome Are Targets of Small RNAs. | A Nickolai Tchurikov | 2022-03-10 | International journal of molecular sciences |
| 36608648 | Sweet splicing. | Maria Carmo-Fonseca | 2023-01-05 | Cell |
| 19893625 | Autogenous translational regulation of the Borna disease virus negative control factor X from polycistronic mRNA using host RNA helicases. | Yohei Watanabe | 2009-11-01 | PLoS pathogens |
| 33052334 | Systematic discovery and functional interrogation of SARS-CoV-2 viral RNA-host protein interactions during infection. | A Ryan Flynn | 2020-10-06 | bioRxiv : the preprint server for biology |
| 35283818 | Contribution of DEAD-Box RNA Helicase 21 to the Nucleolar Localization of Porcine Circovirus Type 4 Capsid Protein. | Jianwei Zhou | 2022-01-01 | Frontiers in microbiology |
| 28472472 | Human DDX21 binds and unwinds RNA guanine quadruplexes. | S K Ewan McRae | 2017-06-20 | Nucleic acids research |
| 35271815 | The expanding universe of PARP1-mediated molecular and therapeutic mechanisms. | Dan Huang | 2022-06-16 | Molecular cell |
| 33023047 | Roles of the Non-Structural Proteins of Influenza A Virus. | Wenzhuo Hao | 2020-10-03 | Pathogens (Basel, Switzerland) |
| 36042885 | Protein purification, crystallization, and structure determination of human DEAD-box RNA helicase DDX21 in different unwinding states. | Zijun Chen | 2022-09-16 | STAR protocols |
| 37072811 | LINC00240 in the 6p22.1 risk locus promotes gastric cancer progression through USP10-mediated DDX21 stabilization. | Nasha Zhang | 2023-04-18 | Journal of experimental & clinical cancer research : CR |
| 32098424 | The Central Role of Non-Structural Protein 1 (NS1) in Influenza Biology and Infection. | Nícia Rosário-Ferreira | 2020-02-22 | International journal of molecular sciences |
| 32714761 | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation. | Zijun Chen | 2020-07-01 | Advanced science (Weinheim, Baden-Wurttemberg, Germany) |
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| DDX21-204 | ENST00000685106 | 3152 | 729aa | ENSP00000510328 |
| DDX21-208 | ENST00000687162 | 2295 | No protein | - |
| DDX21-203 | ENST00000684824 | 4666 | 808aa | ENSP00000508611 |
| DDX21-201 | ENST00000354185 | 4664 | 783aa | ENSP00000346120 |
| DDX21-207 | ENST00000686528 | 3228 | 768aa | ENSP00000508639 |
| DDX21-211 | ENST00000690650 | 3324 | 661aa | ENSP00000508484 |
| DDX21-210 | ENST00000690316 | 4542 | 715aa | ENSP00000510035 |
| DDX21-202 | ENST00000620315 | 4776 | 715aa | ENSP00000480334 |
| DDX21-205 | ENST00000685513 | 4628 | No protein | - |
| DDX21-209 | ENST00000688593 | 4034 | No protein | - |
| DDX21-212 | ENST00000692943 | 4228 | No protein | - |
| DDX21-206 | ENST00000686366 | 2014 | No protein | - |
Phenotypes
| ensgID | Trait | pValue | Pubmed ID |
|---|
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
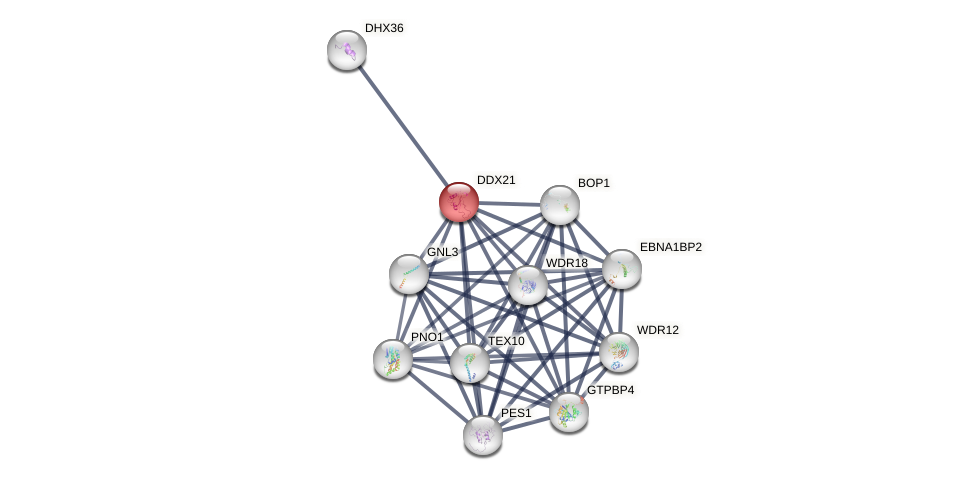
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000165732 | homo_sapiens | ENSP00000379475 | 51.1682 | 27.8037 | homo_sapiens | ENSP00000346120 | 27.9693 | 15.198 |
| ENSG00000165732 | homo_sapiens | ENSP00000385536 | 34.1564 | 20.7133 | homo_sapiens | ENSP00000346120 | 31.8008 | 19.2848 |
| ENSG00000165732 | homo_sapiens | ENSP00000263239 | 40.2985 | 23.5821 | homo_sapiens | ENSP00000346120 | 34.4828 | 20.1788 |
| ENSG00000165732 | homo_sapiens | ENSP00000497641 | 49.1484 | 30.6569 | homo_sapiens | ENSP00000346120 | 25.7982 | 16.092 |
| ENSG00000165732 | homo_sapiens | ENSP00000482680 | 35.9477 | 19.7386 | homo_sapiens | ENSP00000346120 | 35.1213 | 19.2848 |
| ENSG00000165732 | homo_sapiens | ENSP00000293831 | 50 | 31.2808 | homo_sapiens | ENSP00000346120 | 25.9259 | 16.2197 |
| ENSG00000165732 | homo_sapiens | ENSP00000306117 | 43.0962 | 23.8494 | homo_sapiens | ENSP00000346120 | 26.3091 | 14.5594 |
| ENSG00000165732 | homo_sapiens | ENSP00000288071 | 43.215 | 24.2171 | homo_sapiens | ENSP00000346120 | 26.4368 | 14.8148 |
| ENSG00000165732 | homo_sapiens | ENSP00000481495 | 30.7334 | 17.9278 | homo_sapiens | ENSP00000346120 | 33.7165 | 19.6679 |
| ENSG00000165732 | homo_sapiens | ENSP00000416534 | 27.3256 | 15.7946 | homo_sapiens | ENSP00000346120 | 36.0153 | 20.8174 |
| ENSG00000165732 | homo_sapiens | ENSP00000361232 | 31.2332 | 16.756 | homo_sapiens | ENSP00000346120 | 29.7573 | 15.9642 |
| ENSG00000165732 | homo_sapiens | ENSP00000492643 | 17.4436 | 9.32331 | homo_sapiens | ENSP00000346120 | 29.6296 | 15.8365 |
| ENSG00000165732 | homo_sapiens | ENSP00000238146 | 36.8333 | 21.3333 | homo_sapiens | ENSP00000346120 | 28.2248 | 16.3474 |
| ENSG00000165732 | homo_sapiens | ENSP00000225792 | 41.0423 | 24.9186 | homo_sapiens | ENSP00000346120 | 32.1839 | 19.5402 |
| ENSG00000165732 | homo_sapiens | ENSP00000368667 | 38.5103 | 22.504 | homo_sapiens | ENSP00000346120 | 31.0345 | 18.1354 |
| ENSG00000165732 | homo_sapiens | ENSP00000330460 | 39.58 | 21.0016 | homo_sapiens | ENSP00000346120 | 31.2899 | 16.6028 |
| ENSG00000165732 | homo_sapiens | ENSP00000263576 | 41.2008 | 23.3954 | homo_sapiens | ENSP00000346120 | 25.4151 | 14.4317 |
| ENSG00000165732 | homo_sapiens | ENSP00000380495 | 32.7327 | 17.7177 | homo_sapiens | ENSP00000346120 | 27.8416 | 15.0702 |
| ENSG00000165732 | homo_sapiens | ENSP00000314348 | 29.7143 | 15.6571 | homo_sapiens | ENSP00000346120 | 33.2056 | 17.4968 |
| ENSG00000165732 | homo_sapiens | ENSP00000374574 | 28.5714 | 16.5245 | homo_sapiens | ENSP00000346120 | 34.2273 | 19.7957 |
| ENSG00000165732 | homo_sapiens | ENSP00000358716 | 28.6408 | 15.8981 | homo_sapiens | ENSP00000346120 | 30.1405 | 16.7305 |
| ENSG00000165732 | homo_sapiens | ENSP00000442266 | 41.6149 | 22.9814 | homo_sapiens | ENSP00000346120 | 25.6705 | 14.1762 |
| ENSG00000165732 | homo_sapiens | ENSP00000362687 | 72.4559 | 59.7015 | homo_sapiens | ENSP00000346120 | 68.1992 | 56.1941 |
| ENSG00000165732 | homo_sapiens | ENSP00000479504 | 37.5626 | 21.3689 | homo_sapiens | ENSP00000346120 | 28.7356 | 16.3474 |
| ENSG00000165732 | homo_sapiens | ENSP00000359361 | 39.1975 | 23.7654 | homo_sapiens | ENSP00000346120 | 32.4393 | 19.6679 |
| ENSG00000165732 | homo_sapiens | ENSP00000326381 | 50.3685 | 32.9238 | homo_sapiens | ENSP00000346120 | 26.1814 | 17.1137 |
| ENSG00000165732 | homo_sapiens | ENSP00000258772 | 37.8428 | 21.2066 | homo_sapiens | ENSP00000346120 | 26.4368 | 14.8148 |
| ENSG00000165732 | homo_sapiens | ENSP00000494040 | 39.8792 | 24.1692 | homo_sapiens | ENSP00000346120 | 33.7165 | 20.4342 |
| ENSG00000165732 | homo_sapiens | ENSP00000336725 | 39.5455 | 24.0909 | homo_sapiens | ENSP00000346120 | 33.3333 | 20.3065 |
| ENSG00000165732 | homo_sapiens | ENSP00000247003 | 41.8219 | 25.6729 | homo_sapiens | ENSP00000346120 | 25.7982 | 15.8365 |
| ENSG00000165732 | homo_sapiens | ENSP00000330349 | 39.0675 | 21.865 | homo_sapiens | ENSP00000346120 | 31.0345 | 17.3691 |
| ENSG00000165732 | homo_sapiens | ENSP00000242776 | 50.8197 | 27.4005 | homo_sapiens | ENSP00000346120 | 27.7139 | 14.9425 |
| ENSG00000165732 | homo_sapiens | ENSP00000233084 | 30.2703 | 18.5135 | homo_sapiens | ENSP00000346120 | 28.6079 | 17.4968 |
| ENSG00000165732 | homo_sapiens | ENSP00000304072 | 30.193 | 17.1396 | homo_sapiens | ENSP00000346120 | 33.9719 | 19.2848 |
| ENSG00000165732 | homo_sapiens | ENSP00000350698 | 45.9341 | 27.9121 | homo_sapiens | ENSP00000346120 | 26.6922 | 16.2197 |
| ENSG00000165732 | homo_sapiens | ENSP00000310723 | 31.4634 | 18.0488 | homo_sapiens | ENSP00000346120 | 32.9502 | 18.9017 |
| ENSG00000165732 | homo_sapiens | ENSP00000332340 | 37.037 | 20.5556 | homo_sapiens | ENSP00000346120 | 25.5428 | 14.1762 |
| ENSG00000165732 | homo_sapiens | ENSP00000424838 | 35.7735 | 20.0276 | homo_sapiens | ENSP00000346120 | 33.0779 | 18.5185 |
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 99.3614 | 98.9783 | nomascus_leucogenys | ENSNLEP00000014136 | 99.3614 | 98.9783 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 99.8723 | 99.7446 | pan_paniscus | ENSPPAP00000027204 | 99.8723 | 99.7446 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 99.3614 | 99.2337 | pongo_abelii | ENSPPYP00000002774 | 99.3614 | 99.2337 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 99.8723 | 99.8723 | pan_troglodytes | ENSPTRP00000004431 | 99.8723 | 99.8723 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 99.8723 | 99.6169 | gorilla_gorilla | ENSGGOP00000004968 | 99.8723 | 99.6169 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 80.7152 | 69.8595 | sarcophilus_harrisii | ENSSHAP00000006129 | 73.3179 | 63.4571 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 40.8685 | 37.037 | notamacropus_eugenii | ENSMEUP00000009670 | 52.7183 | 47.7759 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 82.6309 | 78.0332 | choloepus_hoffmanni | ENSCHOP00000000205 | 84.1352 | 79.4538 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 89.3997 | 84.6743 | dasypus_novemcinctus | ENSDNOP00000001301 | 91.3838 | 86.5535 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 76.5006 | 71.5198 | erinaceus_europaeus | ENSEEUP00000005438 | 85.9397 | 80.3443 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 91.6986 | 86.0792 | echinops_telfairi | ENSETEP00000011424 | 91.1168 | 85.533 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 97.8289 | 95.9132 | callithrix_jacchus | ENSCJAP00000023617 | 97.8289 | 95.9132 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 98.7229 | 97.5734 | cercocebus_atys | ENSCATP00000041736 | 98.7229 | 97.5734 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 90.2937 | 89.5275 | macaca_fascicularis | ENSMFAP00000026850 | 98.8811 | 98.042 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 98.8506 | 97.8289 | macaca_mulatta | ENSMMUP00000074286 | 98.8506 | 97.8289 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 98.8506 | 97.7011 | macaca_nemestrina | ENSMNEP00000017716 | 98.8506 | 97.7011 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 98.8506 | 97.7011 | papio_anubis | ENSPANP00000058555 | 98.8506 | 97.7011 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 95.53 | 94.1252 | mandrillus_leucophaeus | ENSMLEP00000009568 | 98.5507 | 97.1014 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 92.9757 | 88.2503 | tursiops_truncatus | ENSTTRP00000015543 | 92.9757 | 88.2503 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 93.4866 | 89.5275 | vulpes_vulpes | ENSVVUP00000027956 | 92.1914 | 88.2872 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 88.1226 | 82.7586 | mus_spretus | MGP_SPRETEiJ_P0024278 | 80.9859 | 76.0563 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 94.1252 | 88.7612 | equus_asinus | ENSEASP00005062054 | 93.8854 | 88.535 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 72.6692 | 66.2835 | capra_hircus | ENSCHIP00000012298 | 77.3098 | 70.5163 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 89.9106 | 86.2069 | capra_hircus | ENSCHIP00000000347 | 93.3687 | 89.5225 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 66.7944 | 61.175 | capra_hircus | ENSCHIP00000002604 | 75.0359 | 68.7231 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 93.2312 | 87.6117 | loxodonta_africana | ENSLAFP00000006679 | 92.7573 | 87.1665 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 93.9974 | 90.166 | panthera_leo | ENSPLOP00000023935 | 94.2382 | 90.3969 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 87.7395 | 83.908 | ailuropoda_melanoleuca | ENSAMEP00000019001 | 93.3424 | 89.2663 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 92.848 | 89.1443 | bos_indicus_hybrid | ENSBIXP00005037028 | 92.7296 | 89.0306 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 93.1034 | 88.1226 | oryctolagus_cuniculus | ENSOCUP00000002509 | 93.4615 | 88.4615 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 94.1252 | 89.0166 | equus_caballus | ENSECAP00000070739 | 93.8854 | 88.7898 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 93.4866 | 89.3997 | canis_lupus_familiaris | ENSCAFP00845004294 | 93.2484 | 89.172 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 93.9974 | 90.2937 | panthera_pardus | ENSPPRP00000021791 | 94.2382 | 90.525 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 93.742 | 89.1443 | marmota_marmota_marmota | ENSMMMP00000010243 | 93.6225 | 89.0306 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 93.2312 | 88.8889 | balaenoptera_musculus | ENSBMSP00010030947 | 92.9936 | 88.6624 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 90.8046 | 85.5683 | cavia_porcellus | ENSCPOP00000013960 | 89.2095 | 84.0652 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 93.9974 | 90.2937 | felis_catus | ENSFCAP00000038652 | 94.2382 | 90.525 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 17.6245 | 16.8582 | ursus_americanus | ENSUAMP00000006470 | 89.6104 | 85.7143 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 69.4764 | 61.3027 | monodelphis_domestica | ENSMODP00000013441 | 81.8045 | 72.1805 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 14.8148 | 13.5377 | bison_bison_bison | ENSBBBP00000015241 | 80.5556 | 73.6111 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 16.2197 | 14.8148 | bison_bison_bison | ENSBBBP00000020269 | 87.5862 | 80 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 92.848 | 89.1443 | bison_bison_bison | ENSBBBP00000027215 | 92.7296 | 89.0306 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 93.1034 | 88.5057 | monodon_monoceros | ENSMMNP00015024484 | 92.8662 | 88.2803 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 92.848 | 88.1226 | sus_scrofa | ENSSSCP00000010925 | 92.7296 | 88.0102 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 86.4623 | 80.7152 | microcebus_murinus | ENSMICP00000020440 | 91.363 | 85.2901 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 94.8914 | 90.0383 | microcebus_murinus | ENSMICP00000013780 | 94.8914 | 90.0383 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 17.1137 | 16.092 | octodon_degus | ENSODEP00000002192 | 93.0556 | 87.5 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 69.7318 | 64.6232 | octodon_degus | ENSODEP00000017469 | 87.0813 | 80.7018 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 87.6117 | 81.9923 | jaculus_jaculus | ENSJJAP00000010473 | 86.8354 | 81.2658 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 92.5926 | 89.0166 | ovis_aries_rambouillet | ENSOARP00020012367 | 92.5926 | 89.0166 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 94.1252 | 90.5492 | ursus_maritimus | ENSUMAP00000010290 | 92.8212 | 89.2947 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 82.5032 | 76.3729 | ochotona_princeps | ENSOPRP00000001518 | 84.2243 | 77.9661 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 93.1034 | 88.378 | delphinapterus_leucas | ENSDLEP00000011886 | 92.8662 | 88.1529 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 93.4866 | 89.272 | physeter_catodon | ENSPCTP00005009053 | 93.2484 | 89.0446 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 87.8672 | 82.6309 | mus_caroli | MGP_CAROLIEiJ_P0023133 | 88.5457 | 83.269 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 88.2503 | 82.7586 | mus_musculus | ENSMUSP00000042691 | 81.1986 | 76.1457 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 85.9515 | 80.5875 | dipodomys_ordii | ENSDORP00000022612 | 86.6152 | 81.2098 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 93.8697 | 90.0383 | mustela_putorius_furo | ENSMPUP00000004568 | 92.803 | 89.0152 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 98.212 | 96.8072 | aotus_nancymaae | ENSANAP00000041650 | 98.212 | 96.8072 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 37.2925 | 36.0153 | saimiri_boliviensis_boliviensis | ENSSBOP00000014827 | 89.2966 | 86.2385 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 81.0983 | 77.6501 | vicugna_pacos | ENSVPAP00000000580 | 93.5199 | 89.5434 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 83.6526 | 78.6718 | tupaia_belangeri | ENSTBEP00000002021 | 85.3977 | 80.3129 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 85.696 | 80.7152 | mesocricetus_auratus | ENSMAUP00000010215 | 84.6154 | 79.6973 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 93.2312 | 88.378 | phocoena_sinus | ENSPSNP00000025950 | 92.9936 | 88.1529 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 92.848 | 88.378 | camelus_dromedarius | ENSCDRP00005025904 | 93.8064 | 89.2903 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 94.8914 | 89.9106 | carlito_syrichta | ENSTSYP00000010355 | 94.7704 | 89.7959 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 93.8697 | 89.9106 | panthera_tigris_altaica | ENSPTIP00000004913 | 94.1101 | 90.1408 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 89.1443 | 84.1635 | rattus_norvegicus | ENSRNOP00000063494 | 89.2583 | 84.2711 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 95.2746 | 90.0383 | prolemur_simus | ENSPSMP00000013091 | 95.2746 | 90.0383 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 87.8672 | 82.2478 | microtus_ochrogaster | ENSMOCP00000024427 | 85.6787 | 80.1992 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 52.6181 | 48.0204 | sorex_araneus | ENSSARP00000012692 | 53.6458 | 48.9583 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 89.1443 | 84.5466 | peromyscus_maniculatus_bairdii | ENSPEMP00000020618 | 89.9485 | 85.3093 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 15.4534 | 14.304 | bos_mutus | ENSBMUP00000002447 | 83.4483 | 77.2414 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 81.8646 | 78.4163 | bos_mutus | ENSBMUP00000030246 | 90.6648 | 86.8458 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 87.8672 | 82.5032 | mus_spicilegus | ENSMSIP00000036566 | 84.7291 | 79.5566 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 87.2286 | 81.7369 | cricetulus_griseus_chok1gshd | ENSCGRP00001015745 | 88.4715 | 82.9016 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 78.5441 | 72.0307 | cricetulus_griseus_chok1gshd | ENSCGRP00001017925 | 83.4464 | 76.5265 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 93.1034 | 90.166 | pteropus_vampyrus | ENSPVAP00000008972 | 93.1034 | 90.166 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 93.1034 | 88.6335 | myotis_lucifugus | ENSMLUP00000021145 | 89.2289 | 84.9449 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 69.4764 | 62.1967 | vombatus_ursinus | ENSVURP00010019615 | 81.194 | 72.6866 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 39.0805 | 35.6322 | rhinolophus_ferrumequinum | ENSRFEP00010010919 | 83.6066 | 76.2295 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 92.848 | 88.2503 | rhinolophus_ferrumequinum | ENSRFEP00010013028 | 92.9668 | 88.3632 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 93.8697 | 90.0383 | neovison_vison | ENSNVIP00000015904 | 92.9204 | 89.1277 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 92.4649 | 88.8889 | bos_taurus | ENSBTAP00000019351 | 92.7017 | 89.1165 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 78.9272 | 75.8621 | heterocephalus_glaber_female | ENSHGLP00000037187 | 95.2234 | 91.5254 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 90.6769 | 89.0166 | rhinopithecus_bieti | ENSRBIP00000023898 | 97.7961 | 96.0055 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 98.7229 | 97.8289 | rhinopithecus_bieti | ENSRBIP00000040622 | 98.7229 | 97.8289 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 17.2414 | 15.7088 | canis_lupus_dingo | ENSCAFP00020027386 | 93.1034 | 84.8276 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 15.7088 | 14.8148 | canis_lupus_dingo | ENSCAFP00020033448 | 88.4892 | 83.4532 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 43.6782 | 41.8902 | canis_lupus_dingo | ENSCAFP00020033452 | 93.956 | 90.1099 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 41.7625 | 40.3576 | sciurus_vulgaris | ENSSVLP00005006489 | 95.614 | 92.3977 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 94.3806 | 90.166 | sciurus_vulgaris | ENSSVLP00005004990 | 94.1401 | 89.9363 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 81.6092 | 70.1149 | phascolarctos_cinereus | ENSPCIP00000021048 | 74.3023 | 63.8372 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 80.5875 | 74.4572 | procavia_capensis | ENSPCAP00000015643 | 91.0534 | 84.127 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 89.7829 | 84.2912 | nannospalax_galili | ENSNGAP00000023849 | 85.3155 | 80.0971 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 92.7203 | 89.0166 | bos_grunniens | ENSBGRP00000017281 | 92.7203 | 89.0166 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 91.1877 | 86.4623 | chinchilla_lanigera | ENSCLAP00000017840 | 92.8479 | 88.0364 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 12.7714 | 11.7497 | cervus_hanglu_yarkandensis | ENSCHYP00000013901 | 60.9756 | 56.0976 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 92.9757 | 89.0166 | cervus_hanglu_yarkandensis | ENSCHYP00000010092 | 92.9757 | 89.0166 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 92.848 | 87.6117 | catagonus_wagneri | ENSCWAP00000028787 | 92.7296 | 87.5 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 93.1034 | 88.7612 | ictidomys_tridecemlineatus | ENSSTOP00000010389 | 92.6302 | 88.31 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 92.2094 | 85.8238 | otolemur_garnettii | ENSOGAP00000001646 | 85.7482 | 79.81 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 70.7535 | 67.433 | otolemur_garnettii | ENSOGAP00000020801 | 91.2685 | 86.9852 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 97.9566 | 96.2963 | cebus_imitator | ENSCCAP00000016971 | 97.9566 | 96.2963 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 93.2312 | 88.5057 | urocitellus_parryii | ENSUPAP00010010699 | 93.9511 | 89.1892 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 92.2094 | 87.8672 | moschus_moschiferus | ENSMMSP00000024491 | 92.0918 | 87.7551 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 92.848 | 88.7612 | moschus_moschiferus | ENSMMSP00000031240 | 92.848 | 88.7612 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 98.7229 | 97.7011 | rhinopithecus_roxellana | ENSRROP00000018691 | 98.7229 | 97.7011 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 87.9949 | 83.014 | mus_pahari | MGP_PahariEiJ_P0086304 | 88.6744 | 83.6551 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 92.3372 | 87.2286 | propithecus_coquereli | ENSPCOP00000015128 | 95.3826 | 90.1055 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 37.5479 | 31.6731 | callorhinchus_milii | ENSCMIP00000045342 | 75.7732 | 63.9175 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 40.8685 | 32.0562 | callorhinchus_milii | ENSCMIP00000045334 | 69.4143 | 54.4469 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 59.7701 | 52.4904 | latimeria_chalumnae | ENSLACP00000000733 | 80.4124 | 70.6186 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 67.0498 | 56.9604 | lepisosteus_oculatus | ENSLOCP00000011126 | 67.5676 | 57.4003 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 69.7318 | 57.0881 | danio_rerio | ENSDARP00000087581 | 69.0266 | 56.5107 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 66.1558 | 54.917 | carassius_auratus | ENSCARP00000065079 | 71.7452 | 59.5568 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 68.8378 | 55.6833 | astyanax_mexicanus | ENSAMXP00000030407 | 68.75 | 55.6122 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 66.9221 | 54.5338 | ictalurus_punctatus | ENSIPUP00000010279 | 68.4967 | 55.817 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 67.433 | 56.3218 | esox_lucius | ENSELUP00000070531 | 68.9295 | 57.5718 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 66.9221 | 56.1941 | electrophorus_electricus | ENSEEEP00000013750 | 69.0382 | 57.971 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 65.5172 | 54.6616 | oncorhynchus_kisutch | ENSOKIP00005075275 | 68.5829 | 57.2192 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 66.6667 | 55.6833 | oncorhynchus_kisutch | ENSOKIP00005117632 | 66.6667 | 55.6833 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 65.9004 | 54.4061 | oncorhynchus_mykiss | ENSOMYP00000088817 | 68.8919 | 56.8758 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 65.7727 | 54.7893 | oncorhynchus_mykiss | ENSOMYP00000083132 | 70.1635 | 58.4469 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 66.2835 | 55.3001 | salmo_salar | ENSSSAP00000062891 | 69.6644 | 58.1208 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 66.2835 | 55.1724 | salmo_salar | ENSSSAP00000062701 | 66.9677 | 55.7419 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 66.1558 | 55.0447 | salmo_trutta | ENSSTUP00000109112 | 70 | 58.2432 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 66.4112 | 55.4278 | salmo_trutta | ENSSTUP00000116107 | 70.0809 | 58.4906 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 64.2401 | 52.4904 | gadus_morhua | ENSGMOP00000027823 | 66.1842 | 54.0789 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 66.6667 | 55.0447 | fundulus_heteroclitus | ENSFHEP00000032538 | 67.4419 | 55.6848 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 64.751 | 53.5121 | xiphophorus_maculatus | ENSXMAP00000031402 | 68.0537 | 56.2416 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 60.6641 | 51.5964 | oryzias_latipes | ENSORLP00000012694 | 73.1895 | 62.2496 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 64.8787 | 54.6616 | cyclopterus_lumpus | ENSCLMP00005008102 | 68.8347 | 57.9946 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 66.539 | 55.6833 | oreochromis_niloticus | ENSONIP00000006910 | 64.1626 | 53.6946 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 66.2835 | 55.0447 | haplochromis_burtoni | ENSHBUP00000015739 | 65.201 | 54.1457 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 66.2835 | 55.4278 | astatotilapia_calliptera | ENSACLP00000012079 | 65.1192 | 54.4542 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 65.3895 | 54.5338 | sparus_aurata | ENSSAUP00010058605 | 62.1359 | 51.8204 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 66.539 | 56.3218 | lates_calcarifer | ENSLCAP00010057246 | 69.5594 | 58.8785 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 70.6258 | 60.5364 | chrysemys_picta_bellii | ENSCPBP00000028182 | 79.113 | 67.8112 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 75.3512 | 62.963 | chrysemys_picta_bellii | ENSCPBP00000028186 | 72.5707 | 60.6396 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 70.7535 | 60.0255 | sphenodon_punctatus | ENSSPUP00000005285 | 77.1588 | 65.4596 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 65.1341 | 55.1724 | crocodylus_porosus | ENSCPRP00005008936 | 83.8816 | 71.0526 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 63.9847 | 55.0447 | crocodylus_porosus | ENSCPRP00005008999 | 82.8099 | 71.2397 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 71.7752 | 60.4087 | laticauda_laticaudata | ENSLLTP00000018417 | 80.6313 | 67.8623 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 13.5377 | 11.1111 | notechis_scutatus | ENSNSUP00000001776 | 67.9487 | 55.7692 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 14.1762 | 11.622 | notechis_scutatus | ENSNSUP00000001783 | 67.6829 | 55.4878 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 47.2542 | 37.8033 | notechis_scutatus | ENSNSUP00000020551 | 68.5185 | 54.8148 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 16.2197 | 12.6437 | notechis_scutatus | ENSNSUP00000005424 | 65.8031 | 51.2953 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 71.3921 | 60.1533 | pseudonaja_textilis | ENSPTXP00000025026 | 80.086 | 67.4785 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 73.1801 | 60.1533 | anas_platyrhynchos_platyrhynchos | ENSAPLP00000012871 | 59.8746 | 49.2163 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 71.7752 | 59.387 | gallus_gallus | ENSGALP00010034189 | 78.9326 | 65.309 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 72.5415 | 60.4087 | meleagris_gallopavo | ENSMGAP00000004155 | 80.6818 | 67.1875 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 68.8378 | 57.9821 | serinus_canaria | ENSSCAP00000017242 | 79.4985 | 66.9617 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 71.7752 | 61.3027 | parus_major | ENSPMJP00000018669 | 78.4916 | 67.0391 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 74.7126 | 61.5581 | anolis_carolinensis | ENSACAP00000009711 | 73.957 | 60.9355 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 66.1558 | 55.0447 | neolamprologus_brichardi | ENSNBRP00000007947 | 68.883 | 57.3138 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 72.0307 | 60.7918 | naja_naja | ENSNNAP00000006536 | 80.9182 | 68.2927 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 73.3078 | 61.175 | podarcis_muralis | ENSPMRP00000015831 | 71.2159 | 59.4293 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 69.7318 | 59.2593 | geospiza_fortis | ENSGFOP00000004677 | 80.2941 | 68.2353 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 72.2861 | 61.0473 | taeniopygia_guttata | ENSTGUP00000032280 | 65.2826 | 55.1326 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 67.0498 | 56.5773 | erpetoichthys_calabaricus | ENSECRP00000001999 | 64.6552 | 54.5567 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 18.3908 | 15.8365 | erpetoichthys_calabaricus | ENSECRP00000030268 | 72 | 62 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 37.6756 | 27.9693 | erpetoichthys_calabaricus | ENSECRP00000028969 | 56.2977 | 41.7939 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 30.9068 | 24.1379 | erpetoichthys_calabaricus | ENSECRP00000022988 | 60.9572 | 47.6071 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 66.6667 | 54.2784 | kryptolebias_marmoratus | ENSKMAP00000027984 | 69.2308 | 56.366 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 71.6475 | 59.7701 | salvator_merianae | ENSSMRP00000011311 | 69.5167 | 57.9926 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 64.8787 | 55.5556 | amphilophus_citrinellus | ENSACIP00000008222 | 68.3715 | 58.5464 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 65.5172 | 55.6833 | dicentrarchus_labrax | ENSDLAP00005022501 | 67.1466 | 57.0681 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 65.3895 | 55.0447 | amphiprion_percula | ENSAPEP00000009887 | 67.9947 | 57.2377 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 45.977 | 40.2299 | oryzias_sinensis | ENSOSIP00000029963 | 79.4702 | 69.5364 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 71.9029 | 61.3027 | pelodiscus_sinensis | ENSPSIP00000005111 | 83.7798 | 71.4286 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 72.5415 | 61.6858 | gopherus_evgoodei | ENSGEVP00005022067 | 80.5674 | 68.5106 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 58.493 | 50.0639 | gopherus_evgoodei | ENSGEVP00005022149 | 77.4958 | 66.3283 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 63.9847 | 55.4278 | chelonoidis_abingdonii | ENSCABP00000031386 | 83.9196 | 72.6968 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 73.9464 | 62.963 | chelonoidis_abingdonii | ENSCABP00000031384 | 82.3613 | 70.128 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 67.1775 | 55.811 | seriola_dumerili | ENSSDUP00000029694 | 69.5767 | 57.8042 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 65.7727 | 55.0447 | cottoperca_gobio | ENSCGOP00000026339 | 68.7583 | 57.5434 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 65.9004 | 56.1941 | struthio_camelus_australis | ENSSCUP00000001245 | 86.8687 | 74.0741 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 71.3921 | 58.2375 | anser_brachyrhynchus | ENSABRP00000025201 | 76.6804 | 62.5514 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 61.4304 | 52.3627 | gasterosteus_aculeatus | ENSGACP00000021188 | 72.1139 | 61.4693 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 65.9004 | 53.7676 | cynoglossus_semilaevis | ENSCSEP00000021828 | 67.451 | 55.0327 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 65.5172 | 54.917 | poecilia_formosa | ENSPFOP00000016355 | 64.7727 | 54.2929 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 55.3001 | 47.1264 | myripristis_murdjan | ENSMMDP00005017934 | 68.5127 | 58.3861 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 67.433 | 56.8327 | myripristis_murdjan | ENSMMDP00005055637 | 68.8396 | 58.0183 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 60.0255 | 51.8519 | sander_lucioperca | ENSSLUP00000038741 | 71.3202 | 61.6085 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 71.7752 | 60.6641 | strigops_habroptila | ENSSHBP00005012376 | 79.1549 | 66.9014 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 65.3895 | 55.0447 | amphiprion_ocellaris | ENSAOCP00000018025 | 67.9947 | 57.2377 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 69.6041 | 60.281 | terrapene_carolina_triunguis | ENSTMTP00000008699 | 78.0802 | 67.6218 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 74.9681 | 62.963 | terrapene_carolina_triunguis | ENSTMTP00000008532 | 72.2906 | 60.7143 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 66.539 | 55.6833 | hucho_hucho | ENSHHUP00000020637 | 67.0528 | 56.1133 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 66.2835 | 55.5556 | hucho_hucho | ENSHHUP00000085494 | 66.199 | 55.4847 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 65.2618 | 54.917 | betta_splendens | ENSBSLP00000033887 | 68.7752 | 57.8735 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 62.8353 | 52.235 | pygocentrus_nattereri | ENSPNAP00000009479 | 69.6884 | 57.932 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 66.2835 | 55.9387 | anabas_testudineus | ENSATEP00000032225 | 68.9243 | 58.1673 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 59.6424 | 51.2133 | seriola_lalandi_dorsalis | ENSSLDP00000033550 | 73.4277 | 63.0503 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 65.7727 | 54.6616 | labrus_bergylta | ENSLBEP00000001586 | 67.7632 | 56.3158 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 57.9821 | 50.1916 | pundamilia_nyererei | ENSPNYP00000020592 | 72.9904 | 63.1833 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 65.0064 | 53.2567 | mastacembelus_armatus | ENSMAMP00000029228 | 68.414 | 56.0484 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 66.2835 | 54.7893 | stegastes_partitus | ENSSPAP00000017417 | 68.8329 | 56.8966 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 66.539 | 55.6833 | oncorhynchus_tshawytscha | ENSOTSP00005071751 | 66.285 | 55.4707 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 46.7433 | 38.5696 | acanthochromis_polyacanthus | ENSAPOP00000027614 | 67.156 | 55.4128 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 71.3921 | 59.8978 | aquila_chrysaetos_chrysaetos | ENSACCP00020003308 | 78.8434 | 66.1495 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 67.0498 | 54.6616 | nothobranchius_furzeri | ENSNFUP00015008868 | 66.2879 | 54.0404 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 66.2835 | 53.5121 | cyprinodon_variegatus | ENSCVAP00000015530 | 68.9243 | 55.6441 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 64.4955 | 55.5556 | denticeps_clupeoides | ENSDCDP00000044252 | 69.3681 | 59.7527 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 66.6667 | 56.1941 | larimichthys_crocea | ENSLCRP00005023244 | 63.8923 | 53.8556 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 65.2618 | 55.4278 | takifugu_rubripes | ENSTRUP00000079783 | 68.407 | 58.0991 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 70.4981 | 59.8978 | ficedula_albicollis | ENSFALP00000013597 | 78.0764 | 66.3366 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 64.8787 | 55.0447 | hippocampus_comes | ENSHCOP00000024838 | 65.1282 | 55.2564 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 65.5172 | 54.023 | cyprinus_carpio_carpio | ENSCCRP00000133305 | 71.0526 | 58.5873 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 66.9221 | 55.9387 | cyprinus_carpio_carpio | ENSCCRP00000116854 | 67.5258 | 56.4433 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 70.1149 | 57.599 | coturnix_japonica | ENSCJPP00005003188 | 78.8793 | 64.7989 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 61.175 | 50.7024 | poecilia_latipinna | ENSPLAP00000014206 | 70.0292 | 58.0409 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 63.3461 | 51.8519 | sinocyclocheilus_grahami | ENSSGRP00000063161 | 70.9585 | 58.083 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 59.1315 | 48.7867 | sinocyclocheilus_grahami | ENSSGRP00000084758 | 70.9035 | 58.4992 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 66.4112 | 55.3001 | maylandia_zebra | ENSMZEP00005015473 | 69.0571 | 57.5033 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 59.6424 | 51.0856 | tetraodon_nigroviridis | ENSTNIP00000016800 | 77.1901 | 66.1157 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 67.1775 | 56.0664 | paramormyrops_kingsleyae | ENSPKIP00000035883 | 70.3209 | 58.6898 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 66.2835 | 55.1724 | scleropages_formosus | ENSSFOP00015038167 | 70.23 | 58.4574 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 62.8353 | 45.8493 | leptobrachium_leishanense | ENSLLEP00000034061 | 56.9444 | 41.5509 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 35.1213 | 27.2031 | leptobrachium_leishanense | ENSLLEP00000024835 | 74.7283 | 57.8804 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 60.1533 | 43.4227 | leptobrachium_leishanense | ENSLLEP00000037685 | 63.7348 | 46.0081 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 65.3895 | 53.3844 | scophthalmus_maximus | ENSSMAP00000060457 | 70.5234 | 57.5758 |
| ENSG00000165732 | homo_sapiens | ENSP00000346120 | 65.3895 | 54.1507 | oryzias_melastigma | ENSOMEP00000003620 | 66.0645 | 54.7097 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0001649 | involved_in osteoblast differentiation | 1 | HDA | Homo_sapiens(9606) | Process |
| GO:0002735 | involved_in positive regulation of myeloid dendritic cell cytokine production | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0003723 | enables RNA binding | 2 | HDA,IBA | Homo_sapiens(9606) | Function |
| GO:0003724 | enables RNA helicase activity | 3 | IBA,IDA,IMP | Homo_sapiens(9606) | Function |
| GO:0003725 | enables double-stranded RNA binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005524 | enables ATP binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0005654 | located_in nucleoplasm | 2 | IDA,TAS | Homo_sapiens(9606) | Component |
| GO:0005694 | located_in chromosome | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005730 | is_active_in nucleolus | 3 | IBA,IDA,NAS | Homo_sapiens(9606) | Component |
| GO:0005739 | located_in mitochondrion | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0005829 | located_in cytosol | 1 | ISS | Homo_sapiens(9606) | Component |
| GO:0006338 | involved_in chromatin remodeling | 1 | NAS | Homo_sapiens(9606) | Process |
| GO:0006364 | involved_in rRNA processing | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0006366 | involved_in transcription by RNA polymerase II | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0016020 | located_in membrane | 1 | HDA | Homo_sapiens(9606) | Component |
| GO:0016887 | enables ATP hydrolysis activity | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0019843 | enables rRNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0030515 | enables snoRNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0035066 | involved_in positive regulation of histone acetylation | 1 | NAS | Homo_sapiens(9606) | Process |
| GO:0035198 | enables miRNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0042802 | enables identical protein binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0043123 | involved_in positive regulation of I-kappaB kinase/NF-kappaB signaling | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0043330 | involved_in response to exogenous dsRNA | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0045087 | involved_in innate immune response | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0045943 | involved_in positive regulation of transcription by RNA polymerase I | 1 | NAS | Homo_sapiens(9606) | Process |
| GO:0045944 | involved_in positive regulation of transcription by RNA polymerase II | 1 | NAS | Homo_sapiens(9606) | Process |
| GO:0045945 | involved_in positive regulation of transcription by RNA polymerase III | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0051607 | involved_in defense response to virus | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0062176 | involved_in R-loop processing | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0097322 | enables 7SK snRNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0110016 | part_of B-WICH complex | 1 | IDA | Homo_sapiens(9606) | Component |