RBP Type
Non-canonical_RBPs
Diseases
SARS-COV-2
Drug
N.A.
Main interacting RNAs
N.A.
Moonlighting functions
Metabolic enzyme
Localizations
N.A.
BulkPerturb-seq
Ensembl ID ENSG00000164904 Gene ID
501 Accession
877
Symbol
ALDH7A1
Alias
EPD;PDE;ATQ1
Full Name
aldehyde dehydrogenase 7 family member A1
Status
Confidence
Length
64163 bases
Strand
Minus strand
Position
5 : 126531200 - 126595362
RNA binding domain
N.A.
Summary
The protein encoded by this gene is a member of subfamily 7 in the aldehyde dehydrogenase gene family. These enzymes are thought to play a major role in the detoxification of aldehydes generated by alcohol metabolism and lipid peroxidation. This particular member has homology to a previously described protein from the green garden pea, the 26g pea turgor protein. It is also involved in lysine catabolism that is known to occur in the mitochondrial matrix. Recent reports show that this protein is found both in the cytosol and the mitochondria, and the two forms likely arise from the use of alternative translation initiation sites. An additional variant encoding a different isoform has also been found for this gene. Mutations in this gene are associated with pyridoxine-dependent epilepsy. Several related pseudogenes have also been identified. [provided by RefSeq, Jan 2011]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
Name
Transcript ID
bp
Protein
Translation ID
ALDH7A1-234
ENST00000637782
1773
521aa
ENSP00000490024
ALDH7A1-216
ENST00000635851
2135
545aa
ENSP00000490819
ALDH7A1-201
ENST00000409134
4765
539aa
ENSP00000387123
ALDH7A1-226
ENST00000636872
2747
97aa
ENSP00000490919
ALDH7A1-231
ENST00000637206
2466
479aa
ENSP00000489895
ALDH7A1-232
ENST00000637272
2554
536aa
ENSP00000489686
ALDH7A1-228
ENST00000636886
2321
472aa
ENSP00000490371
ALDH7A1-227
ENST00000636879
2501
554aa
ENSP00000490811
ALDH7A1-225
ENST00000636808
2639
77aa
ENSP00000490833
ALDH7A1-224
ENST00000636743
2332
499aa
ENSP00000489725
ALDH7A1-215
ENST00000553117
1935
475aa
ENSP00000448593
ALDH7A1-208
ENST00000485852
579
No protein
-
ALDH7A1-209
ENST00000497231
2255
No protein
-
ALDH7A1-237
ENST00000638010
1755
No protein
-
ALDH7A1-223
ENST00000636482
1337
No protein
-
ALDH7A1-205
ENST00000458249
2037
97aa
ENSP00000403929
ALDH7A1-233
ENST00000637292
1222
164aa
ENSP00000490655
ALDH7A1-236
ENST00000638008
1865
77aa
ENSP00000490400
ALDH7A1-222
ENST00000636286
1431
No protein
-
ALDH7A1-221
ENST00000636225
1738
77aa
ENSP00000490797
ALDH7A1-219
ENST00000636062
1809
No protein
-
ALDH7A1-206
ENST00000476328
484
No protein
-
ALDH7A1-210
ENST00000503281
937
41aa
ENSP00000488032
ALDH7A1-235
ENST00000637964
1065
348aa
ENSP00000490291
ALDH7A1-212
ENST00000509459
538
27aa
ENSP00000487998
ALDH7A1-203
ENST00000413020
1126
294aa
ENSP00000487936
ALDH7A1-204
ENST00000433026
615
No protein
-
ALDH7A1-229
ENST00000636892
3328
No protein
-
ALDH7A1-214
ENST00000511266
1812
No protein
-
ALDH7A1-213
ENST00000510111
719
239aa
ENSP00000447388
ALDH7A1-202
ENST00000412186
555
136aa
ENSP00000414536
ALDH7A1-211
ENST00000509270
608
193aa
ENSP00000449318
ALDH7A1-220
ENST00000636190
1357
No protein
-
ALDH7A1-230
ENST00000637070
203
No protein
-
ALDH7A1-207
ENST00000479989
598
No protein
-
ALDH7A1-218
ENST00000635933
3247
No protein
-
ALDH7A1-217
ENST00000635858
887
No protein
-
Pathway ID
Pathway Name
Source
ensgID
Trait
pValue
Pubmed ID
29039 Cholesterol 6.7230000E-005 - 29530 Cholesterol 7.3970000E-005 - 88583 Osteoporosis 2E-9 20072603
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
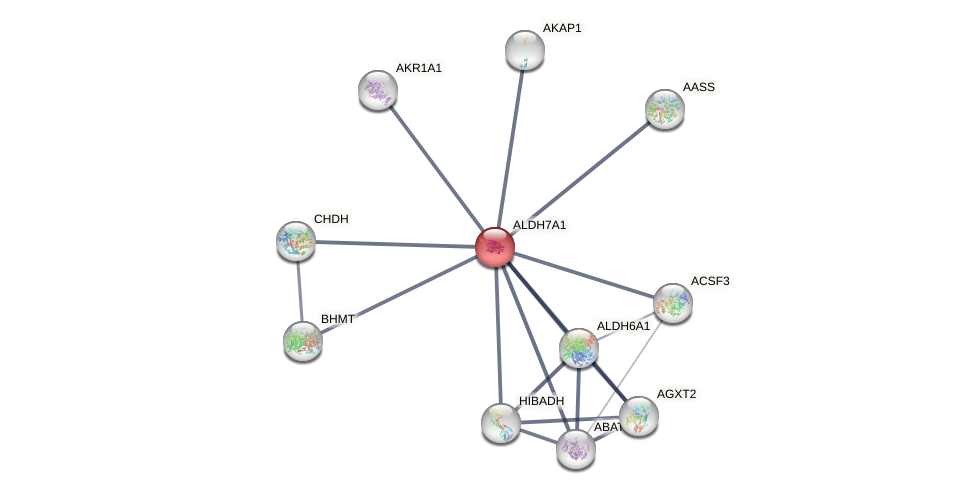
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0004029 enables aldehyde dehydrogenase (NAD+) activity 2 IDA,ISS Homo_sapiens(9606) Function GO:0004043 enables L-aminoadipate-semialdehyde dehydrogenase activity 1 IEA Homo_sapiens(9606) Function GO:0005515 enables protein binding 1 IPI Homo_sapiens(9606) Function GO:0005634 located_in nucleus 1 IEA Homo_sapiens(9606) Component GO:0005739 located_in mitochondrion 1 IDA Homo_sapiens(9606) Component GO:0005759 located_in mitochondrial matrix 1 TAS Homo_sapiens(9606) Component GO:0005829 located_in cytosol 1 IDA Homo_sapiens(9606) Component GO:0006081 involved_in cellular aldehyde metabolic process 1 ISS Homo_sapiens(9606) Process GO:0007605 involved_in sensory perception of sound 1 TAS Homo_sapiens(9606) Process GO:0008802 enables betaine-aldehyde dehydrogenase activity 2 IDA,TAS Homo_sapiens(9606) Function GO:0019285 involved_in glycine betaine biosynthetic process from choline 1 IEA Homo_sapiens(9606) Process GO:0042426 involved_in choline catabolic process 1 TAS Homo_sapiens(9606) Process GO:0042802 enables identical protein binding 1 IDA Homo_sapiens(9606) Function GO:0043878 enables glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity 1 IEA Homo_sapiens(9606) Function GO:0070062 located_in extracellular exosome 1 HDA Homo_sapiens(9606) Component
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.