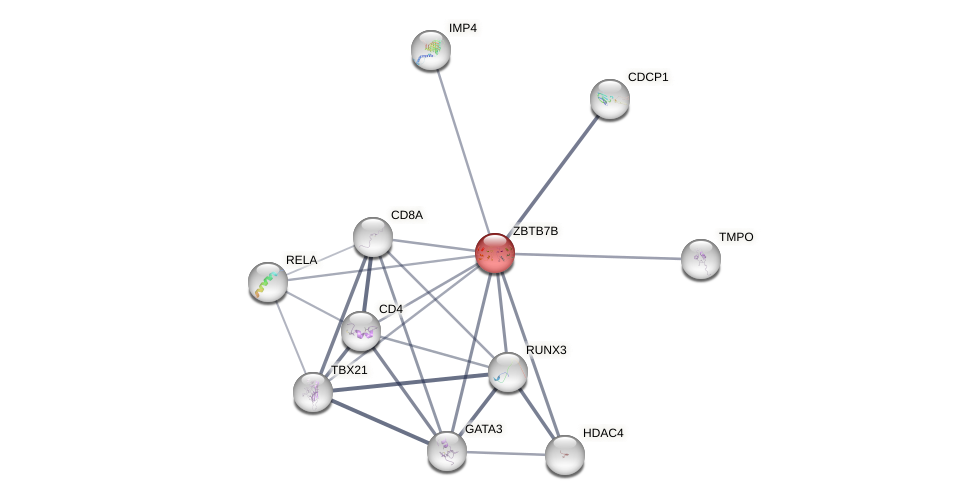
| GO:0000122 | involved_in negative regulation of transcription by RNA polymerase II | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0000978 | enables RNA polymerase II cis-regulatory region sequence-specific DNA binding | 1 | IBA | Homo_sapiens(9606) | Function |
| GO:0000981 | enables DNA-binding transcription factor activity, RNA polymerase II-specific | 1 | IBA | Homo_sapiens(9606) | Function |
| GO:0001217 | enables DNA-binding transcription repressor activity | 1 | ISS | Homo_sapiens(9606) | Function |
| GO:0001228 | enables DNA-binding transcription activator activity, RNA polymerase II-specific | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0001865 | involved_in NK T cell differentiation | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005654 | located_in nucleoplasm | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0006357 | involved_in regulation of transcription by RNA polymerase II | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0006366 | involved_in transcription by RNA polymerase II | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0007398 | involved_in ectoderm development | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0007595 | involved_in lactation | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0010628 | involved_in positive regulation of gene expression | 2 | IBA,ISS | Homo_sapiens(9606) | Process |
| GO:0010629 | involved_in negative regulation of gene expression | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0032740 | involved_in positive regulation of interleukin-17 production | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0032868 | involved_in response to insulin | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0042803 | enables protein homodimerization activity | 1 | ISS | Homo_sapiens(9606) | Function |
| GO:0042826 | enables histone deacetylase binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0043372 | involved_in positive regulation of CD4-positive, alpha-beta T cell differentiation | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0043377 | involved_in negative regulation of CD8-positive, alpha-beta T cell differentiation | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0045944 | involved_in positive regulation of transcription by RNA polymerase II | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0046628 | involved_in positive regulation of insulin receptor signaling pathway | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0046872 | enables metal ion binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0051141 | involved_in negative regulation of NK T cell proliferation | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0090336 | involved_in positive regulation of brown fat cell differentiation | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0120162 | involved_in positive regulation of cold-induced thermogenesis | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:1990837 | enables sequence-specific double-stranded DNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:1990845 | involved_in adaptive thermogenesis | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:2000320 | involved_in negative regulation of T-helper 17 cell differentiation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:2000640 | involved_in positive regulation of SREBP signaling pathway | 1 | ISS | Homo_sapiens(9606) | Process |