| RBP Type |
Non-canonical_RBPs |
| Diseases |
N.A. |
| Drug |
N.A. |
| Main interacting RNAs |
N.A. |
| Moonlighting functions |
E3 ligase |
| Localizations |
N.A. |
|
BulkPerturb-seq
|
|
| Ensembl ID | ENSG00000155827 | Gene ID |
56254 |
Accession |
10062 |
| Symbol |
RNF20 |
Alias |
|
Full Name |
|
| Status |
Confidence |
Length |
29492 bases |
Strand |
Plus strand |
| Position |
9 : 101533853 - 101563344 |
RNA binding domain |
N.A. |
| Summary |
|
RNA binding domains (RBDs)

| Protein |
Domain |
Pfam ID |
E-value |
Domain number |
RNA binding proteomes (RBPomes)
| Pubmed ID |
Full Name |
Cell |
Author |
Time |
Doi |
Literatures on RNA binding capacity
| Pubmed ID |
Title |
Author |
Time |
Journal |
| 26687599 |
Activation of ULK Kinase and Autophagy by GABARAP Trafficking from the Centrosome Is Regulated by WAC and GM130. |
Justin Joachim |
2015-12-17 |
Molecular cell |
| Name |
Transcript ID |
bp |
Protein |
Translation ID |
| RNF20-202 |
ENST00000389120 |
3938 |
975aa |
ENSP00000373772 |
| RNF20-201 |
ENST00000374819 |
596 |
181aa |
ENSP00000363952 |
| RNF20-205 |
ENST00000479306 |
387 |
99aa |
ENSP00000419201 |
| RNF20-206 |
ENST00000481046 |
601 |
No protein |
- |
| RNF20-203 |
ENST00000466817 |
784 |
145aa |
ENSP00000418924 |
| RNF20-204 |
ENST00000478072 |
609 |
No protein |
- |
| Pathway ID |
Pathway Name |
Source |
| | |
| ensgID |
Trait |
pValue |
Pubmed ID |
| ensgID | SNP | Chromosome | Position |
Trait | PubmedID | Or or BEAT |
EFO ID |
|---|
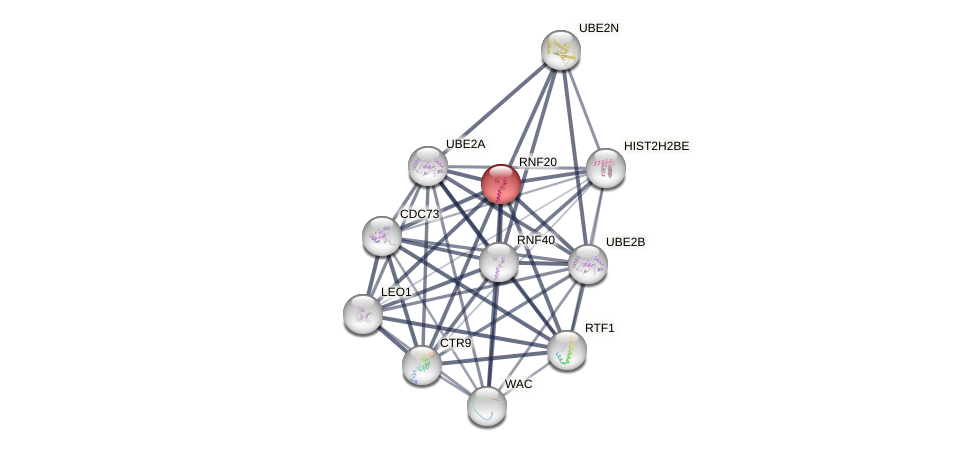
Protein-Protein Interaction (PPI)
| Ensembl ID |
Source |
Target |
| Species |
Protein ID |
Perc_pos |
Perc_id |
Species |
Protein ID |
Perc_pos |
Perc_id |
| Ensembl ID |
Source |
Target |
| Species |
Protein ID |
Perc_pos |
Perc_id |
Species |
Protein ID |
Perc_pos |
Perc_id |
| Go ID |
Go term |
No. evidence |
Entries |
Species |
Category |
| GO:0000151 | part_of ubiquitin ligase complex | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0000209 | involved_in protein polyubiquitination | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0002039 | enables p53 binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0003682 | enables chromatin binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0003713 | enables transcription coactivator activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0003730 | enables mRNA 3-UTR binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0004842 | contributes_to ubiquitin-protein transferase activity | 2 | IDA | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005634 | is_active_in nucleus | 2 | IBA,IDA | Homo_sapiens(9606) | Component |
| GO:0005654 | located_in nucleoplasm | 2 | IDA,TAS | Homo_sapiens(9606) | Component |
| GO:0005730 | located_in nucleolus | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0006338 | involved_in chromatin remodeling | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0006355 | involved_in regulation of DNA-templated transcription | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0006511 | involved_in ubiquitin-dependent protein catabolic process | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0007346 | NOT involved_in regulation of mitotic cell cycle | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0030336 | involved_in negative regulation of cell migration | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0031625 | enables ubiquitin protein ligase binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0033503 | part_of HULC complex | 2 | IBA,IDA | Homo_sapiens(9606) | Component |
| GO:0042393 | enables histone binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0042802 | enables identical protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0045893 | involved_in positive regulation of DNA-templated transcription | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0045944 | involved_in positive regulation of transcription by RNA polymerase II | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0046872 | enables metal ion binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0061630 | enables ubiquitin protein ligase activity | 1 | IBA | Homo_sapiens(9606) | Function |
| GO:0071894 | involved_in histone H2B conserved C-terminal lysine ubiquitination | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0140850 | enables histone H2B C-terminal K residue ubiquitin ligase activity | 1 | IMP | Homo_sapiens(9606) | Function |