RBP Type
Non-canonical_RBPs
Diseases
Drug
N.A.
Main interacting RNAs
N.A.
Moonlighting functions
E3 ligase
Localizations
N.A.
BulkPerturb-seq
Ensembl ID ENSG00000148356 Gene ID
90678 Accession
25135
Symbol
LRSAM1
Alias
TAL;CMT2P;RIFLE
Full Name
leucine rich repeat and sterile alpha motif containing 1
Status
Confidence
Length
52011 bases
Strand
Plus strand
Position
9 : 127451489 - 127503499
RNA binding domain
N.A.
Summary
This gene encodes a ring finger protein involved in a variety of functions, including regulation of signaling pathways and cell adhesion, mediation of self-ubiquitylation, and involvement in cargo sorting during receptor endocytosis. Mutations in this gene have been associated with Charcot-Marie-Tooth disease. Multiple transcript variants encoding different isoforms have been identified for this gene. [provided by RefSeq, Jan 2012]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
27615052 A novel missense mutation of CMT2P alters transcription machinery.
Bo Hu
2016-12-01
Annals of neurology
35842440 C698R mutation in Lrsam1 gene impairs nerve regeneration in a CMT2P mouse model.
Daniel Moiseev
2022-07-16
Scientific reports
Name
Transcript ID
bp
Protein
Translation ID
LRSAM1-220
ENST00000675224
3129
653aa
ENSP00000501869
LRSAM1-204
ENST00000373324
3044
696aa
ENSP00000362421
LRSAM1-213
ENST00000674771
3031
420aa
ENSP00000502627
LRSAM1-228
ENST00000675641
3130
420aa
ENSP00000501845
LRSAM1-209
ENST00000498513
3474
236aa
ENSP00000501637
LRSAM1-227
ENST00000675572
3026
690aa
ENSP00000501598
LRSAM1-241
ENST00000676349
3314
173aa
ENSP00000502155
LRSAM1-240
ENST00000676336
2941
407aa
ENSP00000502686
LRSAM1-238
ENST00000676170
3185
750aa
ENSP00000502177
LRSAM1-214
ENST00000674784
2931
254aa
ENSP00000501837
LRSAM1-231
ENST00000675789
2926
663aa
ENSP00000501954
LRSAM1-201
ENST00000300417
3120
723aa
ENSP00000300417
LRSAM1-224
ENST00000675448
3096
723aa
ENSP00000502167
LRSAM1-202
ENST00000323301
3376
723aa
ENSP00000322937
LRSAM1-205
ENST00000472068
2531
85aa
ENSP00000501555
LRSAM1-217
ENST00000675141
3261
690aa
ENSP00000502420
LRSAM1-211
ENST00000674516
3827
638aa
ENSP00000502441
LRSAM1-232
ENST00000675883
3180
696aa
ENSP00000501592
LRSAM1-234
ENST00000676014
2934
704aa
ENSP00000502058
LRSAM1-208
ENST00000486587
985
No protein
-
LRSAM1-207
ENST00000485704
1227
No protein
-
LRSAM1-233
ENST00000675945
3075
420aa
ENSP00000501835
LRSAM1-229
ENST00000675657
3102
432aa
ENSP00000502002
LRSAM1-239
ENST00000676318
5283
420aa
ENSP00000502300
LRSAM1-221
ENST00000675253
3151
428aa
ENSP00000502557
LRSAM1-215
ENST00000674970
3465
165aa
ENSP00000502493
LRSAM1-219
ENST00000675213
2913
708aa
ENSP00000502218
LRSAM1-203
ENST00000373322
2849
723aa
ENSP00000362419
LRSAM1-226
ENST00000675558
767
No protein
-
LRSAM1-223
ENST00000675445
3026
149aa
ENSP00000502253
LRSAM1-212
ENST00000674621
1932
No protein
-
LRSAM1-218
ENST00000675198
2636
No protein
-
LRSAM1-242
ENST00000676399
2663
No protein
-
LRSAM1-230
ENST00000675662
2551
No protein
-
LRSAM1-237
ENST00000676137
2786
No protein
-
LRSAM1-216
ENST00000675012
2703
No protein
-
LRSAM1-210
ENST00000674511
2358
No protein
-
LRSAM1-243
ENST00000676409
2818
No protein
-
LRSAM1-225
ENST00000675521
2666
No protein
-
LRSAM1-236
ENST00000676106
2796
No protein
-
LRSAM1-235
ENST00000676035
2420
No protein
-
LRSAM1-222
ENST00000675364
1364
No protein
-
LRSAM1-206
ENST00000483302
3421
No protein
-
Pathway ID
Pathway Name
Source
ensgID
Trait
pValue
Pubmed ID
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
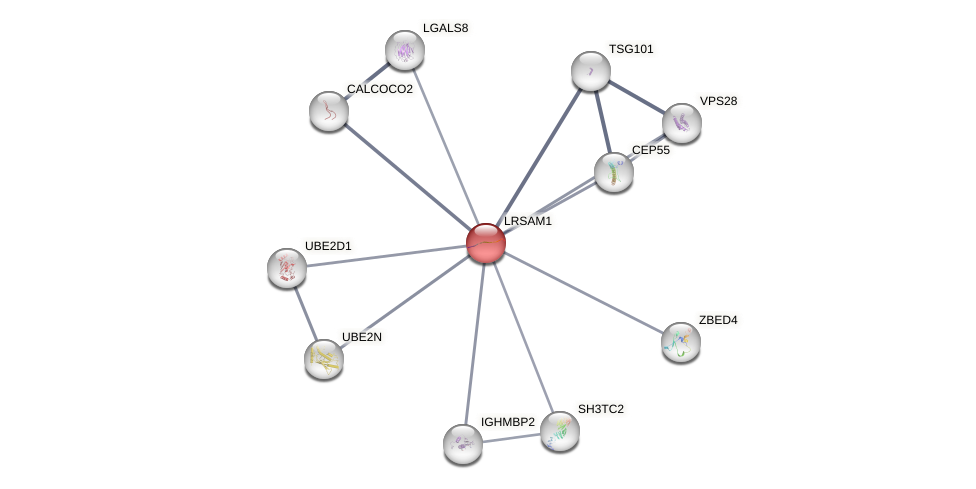
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0000209 involved_in protein polyubiquitination 1 IDA Homo_sapiens(9606) Process GO:0004842 enables ubiquitin-protein transferase activity 1 IDA Homo_sapiens(9606) Function GO:0005515 enables protein binding 1 IPI Homo_sapiens(9606) Function GO:0005737 located_in cytoplasm 1 IDA Homo_sapiens(9606) Component GO:0005829 located_in cytosol 1 IDA Homo_sapiens(9606) Component GO:0006914 involved_in autophagy 1 IEA Homo_sapiens(9606) Process GO:0016020 located_in membrane 1 IDA Homo_sapiens(9606) Component GO:0030163 involved_in protein catabolic process 1 IMP Homo_sapiens(9606) Process GO:0045806 involved_in negative regulation of endocytosis 1 IMP Homo_sapiens(9606) Process GO:0046755 involved_in viral budding 1 IMP Homo_sapiens(9606) Process GO:0046872 enables metal ion binding 1 IEA Homo_sapiens(9606) Function GO:0051865 involved_in protein autoubiquitination 1 IDA Homo_sapiens(9606) Process GO:0061630 enables ubiquitin protein ligase activity 1 IDA Homo_sapiens(9606) Function GO:0070086 involved_in ubiquitin-dependent endocytosis 1 IDA Homo_sapiens(9606) Process GO:1904417 involved_in positive regulation of xenophagy 1 IMP Homo_sapiens(9606) Process GO:2000786 involved_in positive regulation of autophagosome assembly 1 IMP Homo_sapiens(9606) Process
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.