| GO:0001666 | involved_in response to hypoxia | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0003824 | enables catalytic activity | 1 | TAS | Homo_sapiens(9606) | Function |
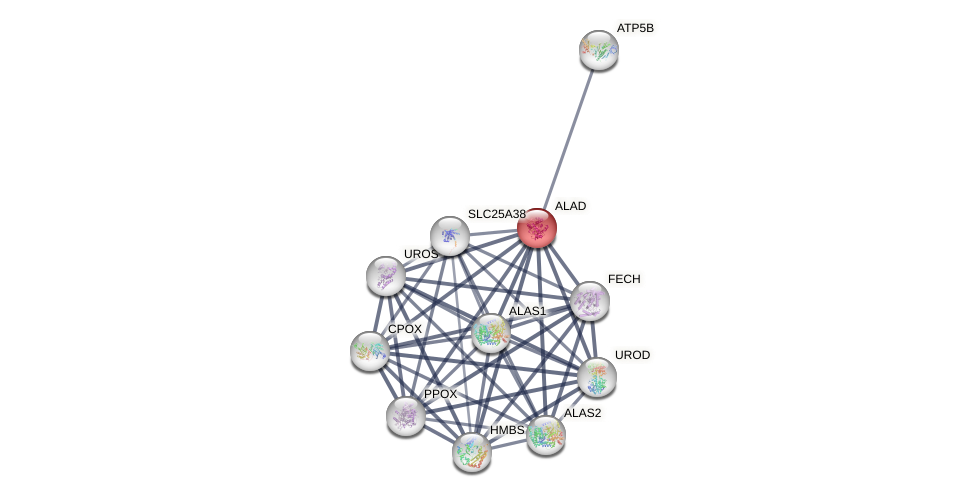
| GO:0004655 | enables porphobilinogen synthase activity | 2 | IBA,IDA | Homo_sapiens(9606) | Function |
| GO:0005576 | located_in extracellular region | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0005634 | located_in nucleus | 1 | HDA | Homo_sapiens(9606) | Component |
| GO:0005829 | is_active_in cytosol | 2 | IBA,TAS | Homo_sapiens(9606) | Component |
| GO:0006782 | involved_in protoporphyrinogen IX biosynthetic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0006783 | involved_in heme biosynthetic process | 2 | IBA,IDA | Homo_sapiens(9606) | Process |
| GO:0006979 | involved_in response to oxidative stress | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0008270 | enables zinc ion binding | 2 | IBA,IDA | Homo_sapiens(9606) | Function |
| GO:0009410 | involved_in response to xenobiotic stimulus | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0009635 | involved_in response to herbicide | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0010039 | involved_in response to iron ion | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0010043 | involved_in response to zinc ion | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0010044 | involved_in response to aluminum ion | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0010212 | involved_in response to ionizing radiation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0010266 | involved_in response to vitamin B1 | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0010269 | involved_in response to selenium ion | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0014823 | involved_in response to activity | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0032025 | involved_in response to cobalt ion | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0032496 | involved_in response to lipopolysaccharide | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0033197 | involved_in response to vitamin E | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0034774 | located_in secretory granule lumen | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0042802 | enables identical protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0043200 | involved_in response to amino acid | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0045471 | involved_in response to ethanol | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0046685 | involved_in response to arsenic-containing substance | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0046686 | involved_in response to cadmium ion | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0046689 | involved_in response to mercury ion | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0051260 | involved_in protein homooligomerization | 1 | IPI | Homo_sapiens(9606) | Process |
| GO:0051384 | involved_in response to glucocorticoid | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0051597 | involved_in response to methylmercury | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0070062 | located_in extracellular exosome | 1 | HDA | Homo_sapiens(9606) | Component |
| GO:0070541 | involved_in response to platinum ion | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0070542 | involved_in response to fatty acid | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0071284 | involved_in cellular response to lead ion | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0071353 | involved_in cellular response to interleukin-4 | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:1901799 | involved_in negative regulation of proteasomal protein catabolic process | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:1904813 | located_in ficolin-1-rich granule lumen | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:1904854 | enables proteasome core complex binding | 1 | IEA | Homo_sapiens(9606) | Function |