RBP Type
Canonical_RBPs
Diseases
CHIKV SARS-COV-OC43 Flaviviruses HIV RV SARS-COV-2 SINV Dengue Zika
Drug
N.A.
Main interacting RNAs
ncRNA
Moonlighting functions
TF
Localizations
Nucleoli Fibrillar center
BulkPerturb-seq
Ensembl ID ENSG00000147140 Gene ID
4841 Accession
7871
Symbol
NONO
Alias
P54;NMT55;NRB54;MRXS34;P54NRB;PPP1R114
Full Name
non-POU domain containing octamer binding
Status
Confidence
Length
46709 bases
Strand
Plus strand
Position
X : 71254814 - 71301522
RNA binding domain
RRM_1
Summary
This gene encodes an RNA-binding protein which plays various roles in the nucleus, including transcriptional regulation and RNA splicing. A rearrangement between this gene and the transcription factor E3 gene has been observed in papillary renal cell carcinoma. Alternatively spliced transcript variants have been described. Pseudogenes exist on Chromosomes 2 and 16. [provided by RefSeq, Feb 2009]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
ENSP00000504193
RRM_1
PF00076 8.4e-31
1
ENSP00000504193
RRM_1
PF00076 8.4e-31
2
ENSP00000503769
RRM_1
PF00076 8.7e-31
1
ENSP00000503769
RRM_1
PF00076 8.7e-31
2
ENSP00000276079
RRM_1
PF00076 1.9e-30
1
ENSP00000276079
RRM_1
PF00076 1.9e-30
2
ENSP00000362947
RRM_1
PF00076 1.9e-30
1
ENSP00000362947
RRM_1
PF00076 1.9e-30
2
ENSP00000406673
RRM_1
PF00076 1.9e-30
1
ENSP00000406673
RRM_1
PF00076 1.9e-30
2
ENSP00000410299
RRM_1
PF00076 1.9e-30
1
ENSP00000410299
RRM_1
PF00076 1.9e-30
2
ENSP00000415777
RRM_1
PF00076 1.9e-30
1
ENSP00000415777
RRM_1
PF00076 1.9e-30
2
ENSP00000503031
RRM_1
PF00076 1.9e-30
1
ENSP00000503031
RRM_1
PF00076 1.9e-30
2
ENSP00000503233
RRM_1
PF00076 1.9e-30
1
ENSP00000503233
RRM_1
PF00076 1.9e-30
2
ENSP00000504314
RRM_1
PF00076 1.9e-30
1
ENSP00000504314
RRM_1
PF00076 1.9e-30
2
ENSP00000504351
RRM_1
PF00076 1.9e-30
1
ENSP00000504351
RRM_1
PF00076 1.9e-30
2
ENSP00000504007
RRM_1
PF00076 1.9e-30
1
ENSP00000504007
RRM_1
PF00076 1.9e-30
2
ENSP00000504665
RRM_1
PF00076 2e-30
1
ENSP00000504665
RRM_1
PF00076 2e-30
2
ENSP00000504263
RRM_1
PF00076 2.3e-30
1
ENSP00000504263
RRM_1
PF00076 2.3e-30
2
ENSP00000362963
RRM_1
PF00076 2.5e-30
1
ENSP00000362963
RRM_1
PF00076 2.5e-30
2
ENSP00000441364
RRM_1
PF00076 2.6e-21
1
ENSP00000441364
RRM_1
PF00076 2.6e-21
2
ENSP00000503920
RRM_1
PF00076 2.6e-21
1
ENSP00000503920
RRM_1
PF00076 2.6e-21
2
ENSP00000413350
RRM_1
PF00076 2.6e-15
1
ENSP00000504090
RRM_1
PF00076 7.6e-15
1
ENSP00000503929
RRM_1
PF00076 4.1e-11
1
ENSP00000409773
RRM_1
PF00076 7.9e-11
1
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
26087192 Long noncoding RNAs and neuroblastoma.
Kumar Gaurav Pandey
2015-07-30
Oncotarget
35547764 mTOR regulates aerobic glycolysis through NEAT1 and nuclear paraspeckle-mediated mechanism in hepatocellular carcinoma.
Hong Zhang
2022-01-01
Theranostics
35458396 The Interplay between Viruses and Host DNA Sensors.
Sandra Huérfano
2022-03-23
Viruses
28250387 Quantitative Proteomic Profiling of Low-Dose Ionizing Radiation Effects in a Human Skin Model.
M Shawna Hengel
2014-07-29
Proteomes
21177649 Fox-3 and PSF interact to activate neural cell-specific alternative splicing.
K Kee Kim
2011-04-01
Nucleic acids research
33127860 PAR-TERRA is the main contributor to telomeric repeat-containing RNA transcripts in normal and cancer mouse cells.
Nikenza Viceconte
2021-01-01
RNA (New York, N.Y.)
24049082 PARP-1 dependent recruitment of the amyotrophic lateral sclerosis-associated protein FUS/TLS to sites of oxidative DNA damage.
L Stuart Rulten
2014-01-01
Nucleic acids research
29535823 Overexpression of p54nrb /NONO induces differential EPHA6 splicing and contributes to castration-resistant prostate cancer growth.
Ryuji Yamamoto
2018-02-13
Oncotarget
34653618 Prostate cancer: Alternatively spliced mRNA transcripts in tumor progression and their uses as therapeutic targets.
Ali Calderon-Aparicio
2021-12-01
The international journal of biochemistry & cell biology
31733171 Multiple interaction nodes define the postreplication repair response to UV-induced DNA damage that is defective in melanomas and correlated with UV signature mutation load.
Sandra Pavey
2020-01-01
Molecular oncology
29773901 Ets-1 promoter-associated noncoding RNA regulates the NONO/ERG/Ets-1 axis to drive gastric cancer progression.
Dan Li
2018-08-01
Oncogene
34904671 Structural basis of dimerization and nucleic acid binding of human DBHS proteins NONO and PSPC1.
J Gavin Knott
2022-01-11
Nucleic acids research
30824709 SFPQ and NONO suppress RNA:DNA-hybrid-related telomere instability.
Eleonora Petti
2019-03-01
Nature communications
32884448 NONO promotes hepatocellular carcinoma progression by enhancing fatty acids biosynthesis through interacting with ACLY mRNA.
Hongda Ding
2020-01-01
Cancer cell international
31250127 Epigenetics and Type 2 Diabetes Risk.
Sangeeta Dhawan
2019-06-27
Current diabetes reports
22941645 PARP activation regulates the RNA-binding protein NONO in the DNA damage response to DNA double-strand breaks.
Jana Krietsch
2012-11-01
Nucleic acids research
35013116 RNA-binding protein p54nrb /NONO potentiates nuclear EGFR-mediated tumorigenesis of triple-negative breast cancer.
Mengqin Shen
2022-01-10
Cell death & disease
29414680 Mealtime Is NONO Speckled: Timing Hepatic Adaptation to Food.
Manon Torres
2018-02-06
Cell metabolism
Name
Transcript ID
bp
Protein
Translation ID
NONO-223
ENST00000677446
2868
471aa
ENSP00000503031
NONO-222
ENST00000677274
3208
471aa
ENSP00000504314
NONO-215
ENST00000535149
2882
382aa
ENSP00000441364
NONO-206
ENST00000420903
2812
471aa
ENSP00000410299
NONO-203
ENST00000373856
1726
515aa
ENSP00000362963
NONO-240
ENST00000679267
4678
No protein
-
NONO-225
ENST00000677766
4895
No protein
-
NONO-211
ENST00000473525
3206
No protein
-
NONO-233
ENST00000678437
2667
468aa
ENSP00000504007
NONO-217
ENST00000676499
3454
No protein
-
NONO-216
ENST00000676495
3909
No protein
-
NONO-230
ENST00000678323
3596
No protein
-
NONO-231
ENST00000678335
2674
360aa
ENSP00000503769
NONO-235
ENST00000678830
2768
501aa
ENSP00000504263
NONO-226
ENST00000677826
3242
No protein
-
NONO-239
ENST00000679254
2438
No protein
-
NONO-221
ENST00000677245
3263
130aa
ENSP00000503929
NONO-227
ENST00000677879
2483
411aa
ENSP00000504090
NONO-202
ENST00000373841
2606
471aa
ENSP00000362947
NONO-220
ENST00000677218
3669
No protein
-
NONO-201
ENST00000276079
2661
471aa
ENSP00000276079
NONO-237
ENST00000679029
2558
360aa
ENSP00000504193
NONO-232
ENST00000678414
987
No protein
-
NONO-218
ENST00000676797
2951
382aa
ENSP00000503920
NONO-238
ENST00000679062
2214
No protein
-
NONO-234
ENST00000678660
2655
476aa
ENSP00000504665
NONO-219
ENST00000677014
2998
56aa
ENSP00000503813
NONO-236
ENST00000678852
1051
No protein
-
NONO-212
ENST00000474431
644
No protein
-
NONO-229
ENST00000678231
2744
471aa
ENSP00000503233
NONO-224
ENST00000677612
2995
471aa
ENSP00000504351
NONO-209
ENST00000471419
3932
No protein
-
NONO-228
ENST00000677977
4330
No protein
-
NONO-210
ENST00000472185
397
No protein
-
NONO-204
ENST00000413858
855
247aa
ENSP00000413350
NONO-213
ENST00000486613
555
No protein
-
NONO-207
ENST00000450092
1710
471aa
ENSP00000415777
NONO-208
ENST00000454976
1520
471aa
ENSP00000406673
NONO-205
ENST00000418921
703
234aa
ENSP00000409773
NONO-214
ENST00000490044
3215
No protein
-
Pathway ID
Pathway Name
Source
ensgID
Trait
pValue
Pubmed ID
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
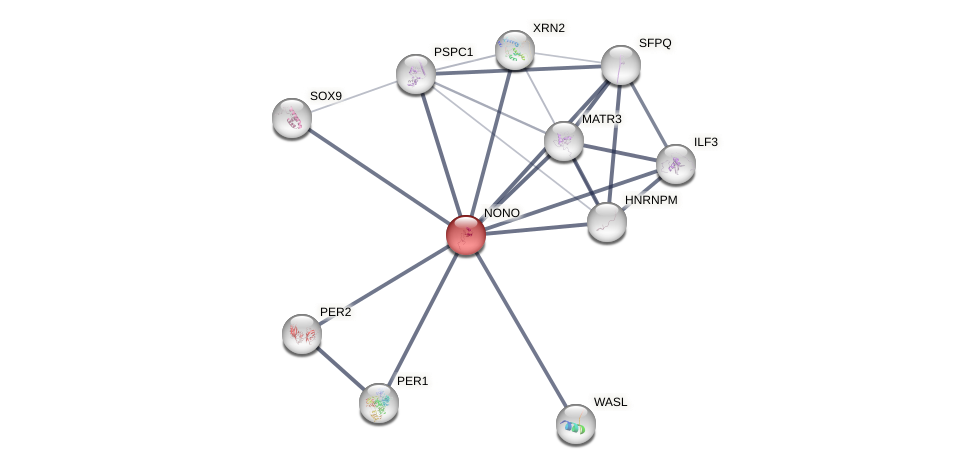
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
ENSG00000147140 homo_sapiens ENSP00000343966 64.8184 54.6845 homo_sapiens ENSP00000276079 71.9745 60.7219 ENSG00000147140 homo_sapiens ENSP00000294428 17.0767 10.9986 homo_sapiens ENSP00000276079 25.0531 16.1359 ENSG00000147140 homo_sapiens ENSP00000454545 15.5056 7.86517 homo_sapiens ENSP00000276079 29.2994 14.862 ENSG00000147140 homo_sapiens ENSP00000364912 6.00437 3.43886 homo_sapiens ENSP00000276079 46.7091 26.7516 ENSG00000147140 homo_sapiens ENSP00000358799 15.0461 7.3695 homo_sapiens ENSP00000276079 31.2102 15.2866 ENSG00000147140 homo_sapiens ENSP00000482277 20.1624 11.7727 homo_sapiens ENSP00000276079 31.6348 18.4713 ENSG00000147140 homo_sapiens ENSP00000349748 48.9392 39.604 homo_sapiens ENSP00000276079 73.4607 59.448
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
ENSG00000147140 homo_sapiens ENSP00000276079 95.7537 95.3291 nomascus_leucogenys ENSNLEP00000040560 97.4082 96.9762 ENSG00000147140 homo_sapiens ENSP00000276079 100 100 pan_paniscus ENSPPAP00000018798 100 100 ENSG00000147140 homo_sapiens ENSP00000276079 100 100 pongo_abelii ENSPPYP00000022896 100 100 ENSG00000147140 homo_sapiens ENSP00000276079 100 100 pan_troglodytes ENSPTRP00000037845 100 100 ENSG00000147140 homo_sapiens ENSP00000276079 88.9597 87.0488 gorilla_gorilla ENSGGOP00000050481 95.0113 92.9705 ENSG00000147140 homo_sapiens ENSP00000276079 79.4055 76.8577 ornithorhynchus_anatinus ENSOANP00000051781 86.5741 83.7963 ENSG00000147140 homo_sapiens ENSP00000276079 94.2675 90.8705 sarcophilus_harrisii ENSSHAP00000011879 93.2773 89.916 ENSG00000147140 homo_sapiens ENSP00000276079 86.8365 83.2272 notamacropus_eugenii ENSMEUP00000003919 86.8365 83.2272 ENSG00000147140 homo_sapiens ENSP00000276079 38.0042 38.0042 choloepus_hoffmanni ENSCHOP00000002068 72.1774 72.1774 ENSG00000147140 homo_sapiens ENSP00000276079 42.2505 39.2781 dasypus_novemcinctus ENSDNOP00000026746 85.4077 79.3991 ENSG00000147140 homo_sapiens ENSP00000276079 97.4522 97.4522 erinaceus_europaeus ENSEEUP00000013151 98.0769 98.0769 ENSG00000147140 homo_sapiens ENSP00000276079 83.0149 82.8026 echinops_telfairi ENSETEP00000011211 84.4492 84.2333 ENSG00000147140 homo_sapiens ENSP00000276079 100 100 callithrix_jacchus ENSCJAP00000024753 100 100 ENSG00000147140 homo_sapiens ENSP00000276079 90.0212 88.9597 cercocebus_atys ENSCATP00000032981 94.2222 93.1111 ENSG00000147140 homo_sapiens ENSP00000276079 93.2059 92.9936 macaca_fascicularis ENSMFAP00000041676 90.8903 90.6832 ENSG00000147140 homo_sapiens ENSP00000276079 93.8429 92.9936 macaca_mulatta ENSMMUP00000040988 87.6984 86.9048 ENSG00000147140 homo_sapiens ENSP00000276079 100 100 macaca_nemestrina ENSMNEP00000002151 100 100 ENSG00000147140 homo_sapiens ENSP00000276079 97.4522 97.4522 papio_anubis ENSPANP00000000606 98.4979 98.4979 ENSG00000147140 homo_sapiens ENSP00000276079 100 100 mandrillus_leucophaeus ENSMLEP00000036757 100 100 ENSG00000147140 homo_sapiens ENSP00000276079 99.3631 99.3631 tursiops_truncatus ENSTTRP00000009104 99.5745 99.5745 ENSG00000147140 homo_sapiens ENSP00000276079 95.966 95.1168 vulpes_vulpes ENSVVUP00000004359 96.1702 95.3191 ENSG00000147140 homo_sapiens ENSP00000276079 99.3631 99.3631 equus_asinus ENSEASP00005052463 99.5745 99.5745 ENSG00000147140 homo_sapiens ENSP00000276079 75.1592 71.9745 capra_hircus ENSCHIP00000005512 86.7647 83.0882 ENSG00000147140 homo_sapiens ENSP00000276079 41.6136 40.552 capra_hircus ENSCHIP00000031269 92.891 90.5213 ENSG00000147140 homo_sapiens ENSP00000276079 98.5138 98.3015 capra_hircus ENSCHIP00000000521 98.7234 98.5106 ENSG00000147140 homo_sapiens ENSP00000276079 98.9384 98.7261 loxodonta_africana ENSLAFP00000011229 98.7288 98.517 ENSG00000147140 homo_sapiens ENSP00000276079 99.3631 99.3631 panthera_leo ENSPLOP00000029653 99.5745 99.5745 ENSG00000147140 homo_sapiens ENSP00000276079 98.5138 98.5138 ailuropoda_melanoleuca ENSAMEP00000016970 94.3089 94.3089 ENSG00000147140 homo_sapiens ENSP00000276079 90.2335 89.8089 bos_indicus_hybrid ENSBIXP00005024976 97.254 96.7963 ENSG00000147140 homo_sapiens ENSP00000276079 99.1507 99.1507 oryctolagus_cuniculus ENSOCUP00000040592 99.3617 99.3617 ENSG00000147140 homo_sapiens ENSP00000276079 99.3631 99.3631 equus_caballus ENSECAP00000036081 93.7876 93.7876 ENSG00000147140 homo_sapiens ENSP00000276079 98.0892 97.8769 canis_lupus_familiaris ENSCAFP00845013151 98.2979 98.0851 ENSG00000147140 homo_sapiens ENSP00000276079 32.0594 31.8471 panthera_pardus ENSPPRP00000020337 96.7949 96.1538 ENSG00000147140 homo_sapiens ENSP00000276079 98.5138 98.5138 balaenoptera_musculus ENSBMSP00010027191 94.3089 94.3089 ENSG00000147140 homo_sapiens ENSP00000276079 77.2824 72.8238 cavia_porcellus ENSCPOP00000018462 88.9976 83.8631 ENSG00000147140 homo_sapiens ENSP00000276079 99.3631 99.3631 felis_catus ENSFCAP00000013735 99.5745 99.5745 ENSG00000147140 homo_sapiens ENSP00000276079 96.8153 96.3907 ursus_americanus ENSUAMP00000009961 97.6445 97.2163 ENSG00000147140 homo_sapiens ENSP00000276079 94.2675 90.4459 monodelphis_domestica ENSMODP00000005475 91.7355 88.0165 ENSG00000147140 homo_sapiens ENSP00000276079 32.2718 31.6348 bison_bison_bison ENSBBBP00000008185 92.1212 90.303 ENSG00000147140 homo_sapiens ENSP00000276079 99.3631 99.3631 monodon_monoceros ENSMMNP00015013757 99.5745 99.5745 ENSG00000147140 homo_sapiens ENSP00000276079 98.3015 98.3015 sus_scrofa ENSSSCP00000056872 65.6738 65.6738 ENSG00000147140 homo_sapiens ENSP00000276079 97.4522 97.4522 microcebus_murinus ENSMICP00000041587 97.6596 97.6596 ENSG00000147140 homo_sapiens ENSP00000276079 91.2951 91.0828 octodon_degus ENSODEP00000003910 94.9227 94.702 ENSG00000147140 homo_sapiens ENSP00000276079 72.8238 67.7282 octodon_degus ENSODEP00000016132 86.398 80.3526 ENSG00000147140 homo_sapiens ENSP00000276079 69.0021 65.1805 octodon_degus ENSODEP00000000634 83.7629 79.1237 ENSG00000147140 homo_sapiens ENSP00000276079 69.6391 65.8174 octodon_degus ENSODEP00000006665 85.1948 80.5195 ENSG00000147140 homo_sapiens ENSP00000276079 94.9045 92.9936 octodon_degus ENSODEP00000002898 95.5128 93.5897 ENSG00000147140 homo_sapiens ENSP00000276079 98.7261 98.3015 jaculus_jaculus ENSJJAP00000014772 98.9362 98.5106 ENSG00000147140 homo_sapiens ENSP00000276079 98.5138 98.3015 ovis_aries_rambouillet ENSOARP00020032000 98.7234 98.5106 ENSG00000147140 homo_sapiens ENSP00000276079 99.3631 99.3631 ursus_maritimus ENSUMAP00000029758 99.5745 99.5745 ENSG00000147140 homo_sapiens ENSP00000276079 57.7495 57.7495 ochotona_princeps ENSOPRP00000001047 66.6667 66.6667 ENSG00000147140 homo_sapiens ENSP00000276079 99.3631 99.3631 delphinapterus_leucas ENSDLEP00000012352 99.5745 99.5745 ENSG00000147140 homo_sapiens ENSP00000276079 98.5138 98.5138 physeter_catodon ENSPCTP00005016156 94.3089 94.3089 ENSG00000147140 homo_sapiens ENSP00000276079 19.5329 19.5329 mus_caroli MGP_CAROLIEiJ_P0092341 91.0891 91.0891 ENSG00000147140 homo_sapiens ENSP00000276079 97.0276 97.0276 mus_musculus ENSMUSP00000033673 96.6173 96.6173 ENSG00000147140 homo_sapiens ENSP00000276079 97.8769 97.6645 dipodomys_ordii ENSDORP00000008733 98.927 98.7124 ENSG00000147140 homo_sapiens ENSP00000276079 99.3631 99.3631 mustela_putorius_furo ENSMPUP00000004898 99.5745 99.5745 ENSG00000147140 homo_sapiens ENSP00000276079 100 100 aotus_nancymaae ENSANAP00000031054 100 100 ENSG00000147140 homo_sapiens ENSP00000276079 96.3907 96.1783 saimiri_boliviensis_boliviensis ENSSBOP00000022320 99.1266 98.9083 ENSG00000147140 homo_sapiens ENSP00000276079 99.3631 99.3631 vicugna_pacos ENSVPAP00000002101 99.5745 99.5745 ENSG00000147140 homo_sapiens ENSP00000276079 62.845 55.6263 chlorocebus_sabaeus ENSCSAP00000010546 76.2887 67.5258 ENSG00000147140 homo_sapiens ENSP00000276079 43.5244 43.3121 tupaia_belangeri ENSTBEP00000002678 49.1607 48.9209 ENSG00000147140 homo_sapiens ENSP00000276079 98.9384 98.7261 mesocricetus_auratus ENSMAUP00000017333 98.3122 98.1013 ENSG00000147140 homo_sapiens ENSP00000276079 99.3631 99.3631 phocoena_sinus ENSPSNP00000025846 99.5745 99.5745 ENSG00000147140 homo_sapiens ENSP00000276079 99.3631 99.3631 camelus_dromedarius ENSCDRP00005025272 99.5745 99.5745 ENSG00000147140 homo_sapiens ENSP00000276079 85.9873 84.7134 carlito_syrichta ENSTSYP00000029441 95.5189 94.1038 ENSG00000147140 homo_sapiens ENSP00000276079 99.1507 99.1507 panthera_tigris_altaica ENSPTIP00000017704 99.3617 99.3617 ENSG00000147140 homo_sapiens ENSP00000276079 97.0276 97.0276 rattus_norvegicus ENSRNOP00000004911 96.0084 96.0084 ENSG00000147140 homo_sapiens ENSP00000276079 99.1507 99.1507 prolemur_simus ENSPSMP00000037262 99.3617 99.3617 ENSG00000147140 homo_sapiens ENSP00000276079 93.4183 92.7813 microtus_ochrogaster ENSMOCP00000005979 97.561 96.8958 ENSG00000147140 homo_sapiens ENSP00000276079 81.9533 78.9809 microtus_ochrogaster ENSMOCP00000023362 88.7356 85.5172 ENSG00000147140 homo_sapiens ENSP00000276079 87.0488 84.7134 microtus_ochrogaster ENSMOCP00000025061 92.9705 90.4762 ENSG00000147140 homo_sapiens ENSP00000276079 91.0828 90.8705 sorex_araneus ENSSARP00000007199 91.863 91.6488 ENSG00000147140 homo_sapiens ENSP00000276079 98.3015 98.3015 peromyscus_maniculatus_bairdii ENSPEMP00000010040 98.0932 98.0932 ENSG00000147140 homo_sapiens ENSP00000276079 98.5138 98.3015 bos_mutus ENSBMUP00000033779 98.7234 98.5106 ENSG00000147140 homo_sapiens ENSP00000276079 32.6964 32.4841 mus_spicilegus ENSMSIP00000012318 92.7711 92.1687 ENSG00000147140 homo_sapiens ENSP00000276079 98.5138 98.3015 cricetulus_griseus_chok1gshd ENSCGRP00001000039 98.3051 98.0932 ENSG00000147140 homo_sapiens ENSP00000276079 98.5138 98.3015 pteropus_vampyrus ENSPVAP00000015573 98.7234 98.5106 ENSG00000147140 homo_sapiens ENSP00000276079 98.9384 98.7261 myotis_lucifugus ENSMLUP00000002266 88.5932 88.403 ENSG00000147140 homo_sapiens ENSP00000276079 94.2675 90.6582 vombatus_ursinus ENSVURP00010016254 93.6709 90.0844 ENSG00000147140 homo_sapiens ENSP00000276079 30.5732 28.8747 rhinolophus_ferrumequinum ENSRFEP00010004471 89.441 84.472 ENSG00000147140 homo_sapiens ENSP00000276079 99.1507 99.1507 neovison_vison ENSNVIP00000012062 99.3617 99.3617 ENSG00000147140 homo_sapiens ENSP00000276079 98.5138 98.3015 bos_taurus ENSBTAP00000016122 98.7234 98.5106 ENSG00000147140 homo_sapiens ENSP00000276079 98.7261 98.5138 heterocephalus_glaber_female ENSHGLP00000035007 87.7358 87.5472 ENSG00000147140 homo_sapiens ENSP00000276079 77.707 77.707 rhinopithecus_bieti ENSRBIP00000028167 100 100 ENSG00000147140 homo_sapiens ENSP00000276079 96.8153 96.3907 canis_lupus_dingo ENSCAFP00020025841 97.6445 97.2163 ENSG00000147140 homo_sapiens ENSP00000276079 99.3631 99.3631 sciurus_vulgaris ENSSVLP00005024969 99.5745 99.5745 ENSG00000147140 homo_sapiens ENSP00000276079 94.2675 90.6582 phascolarctos_cinereus ENSPCIP00000035911 94.2675 90.6582 ENSG00000147140 homo_sapiens ENSP00000276079 99.1507 98.7261 procavia_capensis ENSPCAP00000002386 98.3158 97.8947 ENSG00000147140 homo_sapiens ENSP00000276079 98.3015 98.3015 nannospalax_galili ENSNGAP00000007647 99.3562 99.3562 ENSG00000147140 homo_sapiens ENSP00000276079 87.2611 85.9873 bos_grunniens ENSBGRP00000041223 92.3596 91.0112 ENSG00000147140 homo_sapiens ENSP00000276079 99.5754 99.3631 chinchilla_lanigera ENSCLAP00000005131 98.9451 98.7342 ENSG00000147140 homo_sapiens ENSP00000276079 98.5138 98.3015 cervus_hanglu_yarkandensis ENSCHYP00000036101 98.7234 98.5106 ENSG00000147140 homo_sapiens ENSP00000276079 99.3631 99.3631 catagonus_wagneri ENSCWAP00000004359 99.5745 99.5745 ENSG00000147140 homo_sapiens ENSP00000276079 99.3631 99.3631 ictidomys_tridecemlineatus ENSSTOP00000014206 99.5745 99.5745 ENSG00000147140 homo_sapiens ENSP00000276079 98.3015 98.3015 otolemur_garnettii ENSOGAP00000020270 87.6894 87.6894 ENSG00000147140 homo_sapiens ENSP00000276079 26.1146 25.0531 cebus_imitator ENSCCAP00000016694 86.014 82.5175 ENSG00000147140 homo_sapiens ENSP00000276079 89.172 88.7473 urocitellus_parryii ENSUPAP00010007110 95.4545 95 ENSG00000147140 homo_sapiens ENSP00000276079 82.3779 78.7686 moschus_moschiferus ENSMMSP00000023916 87.5846 83.7472 ENSG00000147140 homo_sapiens ENSP00000276079 99.7877 99.7877 rhinopithecus_roxellana ENSRROP00000039902 99.7877 99.7877 ENSG00000147140 homo_sapiens ENSP00000276079 98.7261 98.7261 mus_pahari MGP_PahariEiJ_P0089955 97.8947 97.8947 ENSG00000147140 homo_sapiens ENSP00000276079 99.3631 99.3631 propithecus_coquereli ENSPCOP00000002715 99.5745 99.5745 ENSG00000147140 homo_sapiens ENSP00000276079 24.4161 11.465 drosophila_melanogaster FBpp0087933 14.5019 6.80958 ENSG00000147140 homo_sapiens ENSP00000276079 51.8047 36.0934 drosophila_melanogaster FBpp0305874 38.7302 26.9841 ENSG00000147140 homo_sapiens ENSP00000276079 53.9278 38.6412 drosophila_melanogaster FBpp0074012 34.2318 24.5283 ENSG00000147140 homo_sapiens ENSP00000276079 22.7176 11.465 ciona_intestinalis ENSCINP00000005147 14.0789 7.10526 ENSG00000147140 homo_sapiens ENSP00000276079 57.9618 43.3121 ciona_intestinalis ENSCINP00000008353 51.2195 38.2739 ENSG00000147140 homo_sapiens ENSP00000276079 39.7028 28.6624 eptatretus_burgeri ENSEBUP00000022862 58.2555 42.0561 ENSG00000147140 homo_sapiens ENSP00000276079 72.6115 60.9342 callorhinchus_milii ENSCMIP00000034826 67.9921 57.0577 ENSG00000147140 homo_sapiens ENSP00000276079 73.0361 66.4544 latimeria_chalumnae ENSLACP00000000670 82.2967 74.8804 ENSG00000147140 homo_sapiens ENSP00000276079 77.2824 67.5159 lepisosteus_oculatus ENSLOCP00000018148 80 69.8901 ENSG00000147140 homo_sapiens ENSP00000276079 72.6115 60.2972 danio_rerio ENSDARP00000014106 77.551 64.3991 ENSG00000147140 homo_sapiens ENSP00000276079 72.8238 60.2972 carassius_auratus ENSCARP00000112808 77.6018 64.2534 ENSG00000147140 homo_sapiens ENSP00000276079 71.9745 60.0849 carassius_auratus ENSCARP00000135067 76.6968 64.0272 ENSG00000147140 homo_sapiens ENSP00000276079 73.673 61.7834 astyanax_mexicanus ENSAMXP00000043977 77.9775 65.3933 ENSG00000147140 homo_sapiens ENSP00000276079 72.3991 60.7219 ictalurus_punctatus ENSIPUP00000026990 78.211 65.5963 ENSG00000147140 homo_sapiens ENSP00000276079 74.31 63.482 esox_lucius ENSELUP00000011074 78.6517 67.191 ENSG00000147140 homo_sapiens ENSP00000276079 73.2484 62.2081 electrophorus_electricus ENSEEEP00000002862 78.0543 66.2896 ENSG00000147140 homo_sapiens ENSP00000276079 74.9469 63.9066 oncorhynchus_kisutch ENSOKIP00005112839 78.9709 67.3378 ENSG00000147140 homo_sapiens ENSP00000276079 72.8238 62.845 oncorhynchus_kisutch ENSOKIP00005112070 77.4266 66.8172 ENSG00000147140 homo_sapiens ENSP00000276079 72.6115 62.4204 oncorhynchus_mykiss ENSOMYP00000099658 77.2009 66.3657 ENSG00000147140 homo_sapiens ENSP00000276079 73.4607 63.2696 oncorhynchus_mykiss ENSOMYP00000048379 77.4049 66.6667 ENSG00000147140 homo_sapiens ENSP00000276079 72.8238 62.4204 salmo_salar ENSSSAP00000095283 77.4266 66.3657 ENSG00000147140 homo_sapiens ENSP00000276079 73.8854 63.0573 salmo_salar ENSSSAP00000142796 74.359 63.4615 ENSG00000147140 homo_sapiens ENSP00000276079 72.8238 62.4204 salmo_trutta ENSSTUP00000007116 77.4266 66.3657 ENSG00000147140 homo_sapiens ENSP00000276079 74.0977 62.6327 salmo_trutta ENSSTUP00000010500 78.0761 65.9955 ENSG00000147140 homo_sapiens ENSP00000276079 47.5584 40.3397 gadus_morhua ENSGMOP00000035692 74.4186 63.1229 ENSG00000147140 homo_sapiens ENSP00000276079 20.1699 15.7113 gadus_morhua ENSGMOP00000051771 59.7484 46.5409 ENSG00000147140 homo_sapiens ENSP00000276079 69.6391 57.1125 fundulus_heteroclitus ENSFHEP00000026714 76.4569 62.704 ENSG00000147140 homo_sapiens ENSP00000276079 72.8238 60.0849 poecilia_reticulata ENSPREP00000014821 77.4266 63.8826 ENSG00000147140 homo_sapiens ENSP00000276079 72.8238 60.0849 xiphophorus_maculatus ENSXMAP00000019931 78.1321 64.4647 ENSG00000147140 homo_sapiens ENSP00000276079 72.1868 58.811 oryzias_latipes ENSORLP00000016028 77.6256 63.242 ENSG00000147140 homo_sapiens ENSP00000276079 73.0361 61.5711 cyclopterus_lumpus ENSCLMP00005000530 77.8281 65.6109 ENSG00000147140 homo_sapiens ENSP00000276079 74.5223 62.6327 oreochromis_niloticus ENSONIP00000025802 79.4118 66.7421 ENSG00000147140 homo_sapiens ENSP00000276079 74.31 62.6327 haplochromis_burtoni ENSHBUP00000009218 79.1855 66.7421 ENSG00000147140 homo_sapiens ENSP00000276079 67.7282 57.3248 astatotilapia_calliptera ENSACLP00000017616 85.0667 72 ENSG00000147140 homo_sapiens ENSP00000276079 21.2314 17.1975 sparus_aurata ENSSAUP00010027204 61.7284 50 ENSG00000147140 homo_sapiens ENSP00000276079 73.2484 61.7834 lates_calcarifer ENSLCAP00010053896 78.2313 65.9864 ENSG00000147140 homo_sapiens ENSP00000276079 87.2611 76.8577 xenopus_tropicalis ENSXETP00000045057 88.3871 77.8495 ENSG00000147140 homo_sapiens ENSP00000276079 89.3843 85.9873 chrysemys_picta_bellii ENSCPBP00000023334 89.5745 86.1702 ENSG00000147140 homo_sapiens ENSP00000276079 74.9469 70.4883 sphenodon_punctatus ENSSPUP00000004038 91.9271 86.4583 ENSG00000147140 homo_sapiens ENSP00000276079 88.9597 84.9257 crocodylus_porosus ENSCPRP00005001943 91.087 86.9565 ENSG00000147140 homo_sapiens ENSP00000276079 83.6518 80.2548 laticauda_laticaudata ENSLLTP00000004029 91.2037 87.5 ENSG00000147140 homo_sapiens ENSP00000276079 74.7346 70.9129 notechis_scutatus ENSNSUP00000019383 91.6667 86.9792 ENSG00000147140 homo_sapiens ENSP00000276079 10.4034 9.97877 pseudonaja_textilis ENSPTXP00000011813 69.0141 66.1972 ENSG00000147140 homo_sapiens ENSP00000276079 81.9533 77.9193 anas_platyrhynchos_platyrhynchos ENSAPLP00000002104 86.7416 82.4719 ENSG00000147140 homo_sapiens ENSP00000276079 89.3843 85.3503 gallus_gallus ENSGALP00010024038 81.9066 78.2101 ENSG00000147140 homo_sapiens ENSP00000276079 89.5966 85.3503 meleagris_gallopavo ENSMGAP00000004030 89.2178 84.9894 ENSG00000147140 homo_sapiens ENSP00000276079 88.535 84.9257 serinus_canaria ENSSCAP00000023613 90.6522 86.9565 ENSG00000147140 homo_sapiens ENSP00000276079 88.535 84.9257 parus_major ENSPMJP00000015144 90.6522 86.9565 ENSG00000147140 homo_sapiens ENSP00000276079 87.2611 83.2272 anolis_carolinensis ENSACAP00000014814 88.1974 84.1202 ENSG00000147140 homo_sapiens ENSP00000276079 74.5223 62.845 neolamprologus_brichardi ENSNBRP00000022276 79.4118 66.9683 ENSG00000147140 homo_sapiens ENSP00000276079 89.5966 85.3503 naja_naja ENSNNAP00000016465 89.9787 85.7143 ENSG00000147140 homo_sapiens ENSP00000276079 89.172 84.7134 podarcis_muralis ENSPMRP00000035190 88.983 84.5339 ENSG00000147140 homo_sapiens ENSP00000276079 84.5011 81.104 geospiza_fortis ENSGFOP00000002481 87.6652 84.141 ENSG00000147140 homo_sapiens ENSP00000276079 88.535 84.7134 taeniopygia_guttata ENSTGUP00000029469 87.7895 84 ENSG00000147140 homo_sapiens ENSP00000276079 78.3439 66.6667 erpetoichthys_calabaricus ENSECRP00000015961 80.3922 68.4096 ENSG00000147140 homo_sapiens ENSP00000276079 73.8854 59.8726 kryptolebias_marmoratus ENSKMAP00000003675 78.5553 63.6569 ENSG00000147140 homo_sapiens ENSP00000276079 89.172 84.2887 salvator_merianae ENSSMRP00000004628 90.7127 85.7451 ENSG00000147140 homo_sapiens ENSP00000276079 22.7176 15.9236 oryzias_javanicus ENSOJAP00000013638 66.0494 46.2963 ENSG00000147140 homo_sapiens ENSP00000276079 74.0977 62.2081 amphilophus_citrinellus ENSACIP00000025785 78.9593 66.2896 ENSG00000147140 homo_sapiens ENSP00000276079 69.8514 58.1741 dicentrarchus_labrax ENSDLAP00005005675 79.2771 66.0241 ENSG00000147140 homo_sapiens ENSP00000276079 67.0913 56.051 amphiprion_percula ENSAPEP00000015979 77.6413 64.8649 ENSG00000147140 homo_sapiens ENSP00000276079 71.9745 58.3864 oryzias_sinensis ENSOSIP00000030424 77.3973 62.7854 ENSG00000147140 homo_sapiens ENSP00000276079 89.172 85.9873 pelodiscus_sinensis ENSPSIP00000011940 91.3043 88.0435 ENSG00000147140 homo_sapiens ENSP00000276079 24.4161 23.7792 gopherus_evgoodei ENSGEVP00005004100 78.7671 76.7123 ENSG00000147140 homo_sapiens ENSP00000276079 26.1146 23.5669 chelonoidis_abingdonii ENSCABP00000008116 82.5503 74.4966 ENSG00000147140 homo_sapiens ENSP00000276079 72.6115 60.9342 seriola_dumerili ENSSDUP00000030376 77.551 65.0794 ENSG00000147140 homo_sapiens ENSP00000276079 23.9915 18.896 cottoperca_gobio ENSCGOP00000021284 60.1064 47.3404 ENSG00000147140 homo_sapiens ENSP00000276079 48.1953 40.9766 cottoperca_gobio ENSCGOP00000021286 77.7397 66.0959 ENSG00000147140 homo_sapiens ENSP00000276079 83.6518 79.4055 struthio_camelus_australis ENSSCUP00000023362 89.7494 85.1936 ENSG00000147140 homo_sapiens ENSP00000276079 88.535 84.7134 anser_brachyrhynchus ENSABRP00000024147 90.2597 86.3636 ENSG00000147140 homo_sapiens ENSP00000276079 73.673 61.9958 gasterosteus_aculeatus ENSGACP00000015633 78.8636 66.3636 ENSG00000147140 homo_sapiens ENSP00000276079 73.8854 61.7834 cynoglossus_semilaevis ENSCSEP00000016649 75.6522 63.2609 ENSG00000147140 homo_sapiens ENSP00000276079 73.0361 61.3588 cynoglossus_semilaevis ENSCSEP00000012659 77.8281 65.3846 ENSG00000147140 homo_sapiens ENSP00000276079 69.0021 55.414 poecilia_formosa ENSPFOP00000016493 77.5656 62.2912 ENSG00000147140 homo_sapiens ENSP00000276079 20.8068 16.7728 myripristis_murdjan ENSMMDP00005001585 47.1154 37.9808 ENSG00000147140 homo_sapiens ENSP00000276079 71.7622 60.5096 sander_lucioperca ENSSLUP00000058768 79.717 67.217 ENSG00000147140 homo_sapiens ENSP00000276079 89.172 85.138 strigops_habroptila ENSSHBP00005021354 88.0503 84.0671 ENSG00000147140 homo_sapiens ENSP00000276079 72.3991 59.6603 amphiprion_ocellaris ENSAOCP00000014238 77.8539 64.1553 ENSG00000147140 homo_sapiens ENSP00000276079 89.3843 85.9873 terrapene_carolina_triunguis ENSTMTP00000020496 89.5745 86.1702 ENSG00000147140 homo_sapiens ENSP00000276079 72.8238 62.6327 hucho_hucho ENSHHUP00000020292 77.4266 66.5914 ENSG00000147140 homo_sapiens ENSP00000276079 74.5223 63.482 hucho_hucho ENSHHUP00000011430 78.3482 66.7411 ENSG00000147140 homo_sapiens ENSP00000276079 73.2484 61.7834 betta_splendens ENSBSLP00000049780 78.2313 65.9864 ENSG00000147140 homo_sapiens ENSP00000276079 73.2484 60.7219 pygocentrus_nattereri ENSPNAP00000023701 69 57.2 ENSG00000147140 homo_sapiens ENSP00000276079 21.8684 17.1975 anabas_testudineus ENSATEP00000052246 66.8831 52.5974 ENSG00000147140 homo_sapiens ENSP00000276079 15.9236 8.06794 ciona_savignyi ENSCSAVP00000018820 21.0674 10.6742 ENSG00000147140 homo_sapiens ENSP00000276079 23.3546 14.0127 ciona_savignyi ENSCSAVP00000008889 16.129 9.67742 ENSG00000147140 homo_sapiens ENSP00000276079 72.6115 60.9342 seriola_lalandi_dorsalis ENSSLDP00000015586 77.551 65.0794 ENSG00000147140 homo_sapiens ENSP00000276079 70.4883 59.6603 labrus_bergylta ENSLBEP00000029908 76.8519 65.0463 ENSG00000147140 homo_sapiens ENSP00000276079 31.6348 26.1146 pundamilia_nyererei ENSPNYP00000024254 87.6471 72.3529 ENSG00000147140 homo_sapiens ENSP00000276079 73.2484 61.3588 mastacembelus_armatus ENSMAMP00000010806 78.4091 65.6818 ENSG00000147140 homo_sapiens ENSP00000276079 73.4607 61.5711 stegastes_partitus ENSSPAP00000016775 78.8155 66.0592 ENSG00000147140 homo_sapiens ENSP00000276079 74.7346 63.6943 oncorhynchus_tshawytscha ENSOTSP00005032570 78.7472 67.1141 ENSG00000147140 homo_sapiens ENSP00000276079 72.8238 62.845 oncorhynchus_tshawytscha ENSOTSP00005036914 77.4266 66.8172 ENSG00000147140 homo_sapiens ENSP00000276079 73.673 63.6943 oncorhynchus_tshawytscha ENSOTSP00005016275 77.8027 67.2646 ENSG00000147140 homo_sapiens ENSP00000276079 72.6115 60.7219 acanthochromis_polyacanthus ENSAPOP00000019832 77.9043 65.1481 ENSG00000147140 homo_sapiens ENSP00000276079 89.3843 85.5626 aquila_chrysaetos_chrysaetos ENSACCP00020020969 91.1255 87.2294 ENSG00000147140 homo_sapiens ENSP00000276079 72.1868 59.8726 cyprinodon_variegatus ENSCVAP00000002834 77.0975 63.9456 ENSG00000147140 homo_sapiens ENSP00000276079 73.4607 61.7834 denticeps_clupeoides ENSDCDP00000032347 77.7528 65.3933 ENSG00000147140 homo_sapiens ENSP00000276079 74.0977 62.4204 larimichthys_crocea ENSLCRP00005027375 79.1383 66.6667 ENSG00000147140 homo_sapiens ENSP00000276079 71.9745 60.5096 takifugu_rubripes ENSTRUP00000013365 76.5237 64.3341 ENSG00000147140 homo_sapiens ENSP00000276079 83.2272 78.3439 ficedula_albicollis ENSFALP00000019115 86.5342 81.457 ENSG00000147140 homo_sapiens ENSP00000276079 73.8854 62.4204 hippocampus_comes ENSHCOP00000025941 82.4645 69.6682 ENSG00000147140 homo_sapiens ENSP00000276079 72.6115 60.0849 cyprinus_carpio_carpio ENSCCRP00000036107 77.3756 64.0272 ENSG00000147140 homo_sapiens ENSP00000276079 72.1868 59.8726 cyprinus_carpio_carpio ENSCCRP00000036098 76.9231 63.8009 ENSG00000147140 homo_sapiens ENSP00000276079 86.1996 81.5287 coturnix_japonica ENSCJPP00005014410 86.383 81.7021 ENSG00000147140 homo_sapiens ENSP00000276079 72.6115 59.2357 poecilia_latipinna ENSPLAP00000013906 77.2009 62.9797 ENSG00000147140 homo_sapiens ENSP00000276079 72.1868 59.2357 sinocyclocheilus_grahami ENSSGRP00000039382 76.9231 63.1222 ENSG00000147140 homo_sapiens ENSP00000276079 71.9745 59.8726 sinocyclocheilus_grahami ENSSGRP00000074039 76.6968 63.8009 ENSG00000147140 homo_sapiens ENSP00000276079 67.9406 57.3248 maylandia_zebra ENSMZEP00005013565 85.3333 72 ENSG00000147140 homo_sapiens ENSP00000276079 70.4883 58.3864 paramormyrops_kingsleyae ENSPKIP00000001034 75.6264 62.6424 ENSG00000147140 homo_sapiens ENSP00000276079 76.8577 65.1805 paramormyrops_kingsleyae ENSPKIP00000035604 80.9843 68.6801 ENSG00000147140 homo_sapiens ENSP00000276079 69.2144 57.3248 scleropages_formosus ENSSFOP00015000148 74.7706 61.9266 ENSG00000147140 homo_sapiens ENSP00000276079 75.5839 64.5435 scleropages_formosus ENSSFOP00015013932 79.6421 68.0089 ENSG00000147140 homo_sapiens ENSP00000276079 84.7134 74.9469 leptobrachium_leishanense ENSLLEP00000037159 80.7692 71.4575 ENSG00000147140 homo_sapiens ENSP00000276079 72.1868 60.2972 scophthalmus_maximus ENSSMAP00000007653 76.9231 64.2534 ENSG00000147140 homo_sapiens ENSP00000276079 71.7622 58.5987 oryzias_melastigma ENSOMEP00000011400 77.169 63.0137
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0001650 located_in fibrillar center 1 IDA Homo_sapiens(9606) Component GO:0002218 involved_in activation of innate immune response 1 IDA Homo_sapiens(9606) Process GO:0003677 enables DNA binding 1 IEA Homo_sapiens(9606) Function GO:0003682 enables chromatin binding 1 ISS Homo_sapiens(9606) Function GO:0003723 enables RNA binding 2 HDA,IBA Homo_sapiens(9606) Function GO:0005515 enables protein binding 1 IPI Homo_sapiens(9606) Function GO:0005634 is_active_in nucleus 3 IBA,IDA,ISS Homo_sapiens(9606) Component GO:0005654 located_in nucleoplasm 1 IDA Homo_sapiens(9606) Component GO:0005694 located_in chromosome 1 IEA Homo_sapiens(9606) Component GO:0006281 involved_in DNA repair 1 IEA Homo_sapiens(9606) Process GO:0006310 involved_in DNA recombination 1 IEA Homo_sapiens(9606) Process GO:0006355 involved_in regulation of DNA-templated transcription 1 IBA Homo_sapiens(9606) Process GO:0006397 involved_in mRNA processing 1 IEA Homo_sapiens(9606) Process GO:0007623 involved_in circadian rhythm 1 ISS Homo_sapiens(9606) Process GO:0008380 involved_in RNA splicing 1 IEA Homo_sapiens(9606) Process GO:0016020 located_in membrane 1 HDA Homo_sapiens(9606) Component GO:0016363 located_in nuclear matrix 1 IDA Homo_sapiens(9606) Component GO:0016607 located_in nuclear speck 1 IEA Homo_sapiens(9606) Component GO:0042382 located_in paraspeckles 1 IDA Homo_sapiens(9606) Component GO:0042752 involved_in regulation of circadian rhythm 1 ISS Homo_sapiens(9606) Process GO:0042802 enables identical protein binding 1 IPI Homo_sapiens(9606) Function GO:0045087 involved_in innate immune response 1 IEA Homo_sapiens(9606) Process GO:0045892 involved_in negative regulation of DNA-templated transcription 1 ISS Homo_sapiens(9606) Process GO:0090575 part_of RNA polymerase II transcription regulator complex 1 ISS Homo_sapiens(9606) Component GO:0090650 involved_in cellular response to oxygen-glucose deprivation 1 IEA Homo_sapiens(9606) Process GO:0106222 enables lncRNA binding 1 IEA Homo_sapiens(9606) Function GO:1903377 involved_in negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway 1 IDA Homo_sapiens(9606) Process GO:1904385 involved_in cellular response to angiotensin 1 IEA Homo_sapiens(9606) Process
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.