RBP Type
Canonical_RBPs
Diseases
N.A.
Drug
N.A.
Main interacting RNAs
N.A.
Moonlighting functions
E3 ligase
Localizations
Nucleoli
BulkPerturb-seq
Ensembl ID ENSG00000146063 Gene ID
90933 Accession
19013
Symbol
TRIM41
Alias
RINCK
Full Name
tripartite motif containing 41
Status
Confidence
Length
13310 bases
Strand
Plus strand
Position
5 : 181222499 - 181235808
RNA binding domain
SPRY
Summary
This gene encodes a member of the tripartite motif (TRIM) family. The TRIM family is characterized by a signature motif composed of a RING finger, one or more B-box domains, and a coiled-coil region. This encoded protein may play a role in protein kinase C signaling. Multiple transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Oct 2011]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
ENSP00000320869
SPRY
PF00622 9.1e-12
1
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
35562987 The Roles of Ubiquitination in Pathogenesis of Influenza Virus Infection.
Eun-Sook Park
2022-04-21
International journal of molecular sciences
34835115 How Influenza A Virus NS1 Deals with the Ubiquitin System to Evade Innate Immunity.
Laurie-Anne Lamotte
2021-11-19
Viruses
Name
Transcript ID
bp
Protein
Translation ID
TRIM41-208
ENST00000515499
914
191aa
ENSP00000426803
TRIM41-201
ENST00000315073
3645
630aa
ENSP00000320869
TRIM41-204
ENST00000508930
3834
No protein
-
TRIM41-202
ENST00000351937
2696
518aa
ENSP00000336749
TRIM41-203
ENST00000503114
1904
362aa
ENSP00000424979
TRIM41-207
ENST00000515223
5707
No protein
-
TRIM41-209
ENST00000515834
7015
No protein
-
TRIM41-206
ENST00000514219
459
116aa
ENSP00000426790
TRIM41-205
ENST00000510072
752
No protein
-
Pathway ID
Pathway Name
Source
ensgID
Trait
pValue
Pubmed ID
36196 Platelet Function Tests 1.4800000E-006 - 36221 Platelet Function Tests 2.1200000E-007 -
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
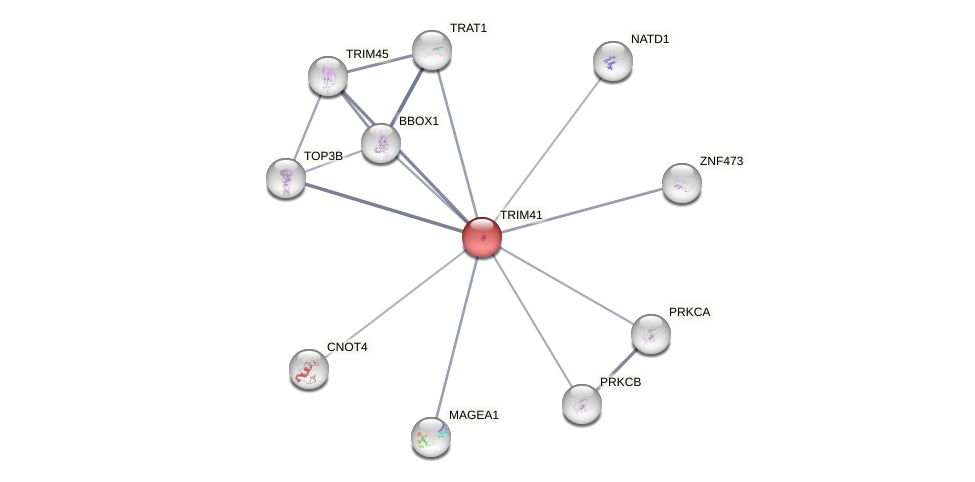
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0005515 enables protein binding 1 IPI Homo_sapiens(9606) Function GO:0005730 located_in nucleolus 1 IDA Homo_sapiens(9606) Component GO:0005737 located_in cytoplasm 1 IEA Homo_sapiens(9606) Component GO:0006513 involved_in protein monoubiquitination 1 IDA Homo_sapiens(9606) Process GO:0008270 enables zinc ion binding 1 IEA Homo_sapiens(9606) Function GO:0016567 involved_in protein ubiquitination 1 IBA Homo_sapiens(9606) Process GO:0016604 is_active_in nuclear body 2 IBA,IDA Homo_sapiens(9606) Component GO:0042802 enables identical protein binding 1 IPI Homo_sapiens(9606) Function GO:0051607 involved_in defense response to virus 1 IDA Homo_sapiens(9606) Process GO:0060340 involved_in positive regulation of type I interferon-mediated signaling pathway 1 IDA Homo_sapiens(9606) Process GO:0061630 enables ubiquitin protein ligase activity 2 IBA,IDA Homo_sapiens(9606) Function GO:0071222 involved_in cellular response to lipopolysaccharide 1 IMP Homo_sapiens(9606) Process GO:0071225 involved_in cellular response to muramyl dipeptide 1 IMP Homo_sapiens(9606) Process
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.