RBP Type
Non-canonical_RBPs
Diseases
N.A.
Drug
N.A.
Main interacting RNAs
N.A.
Moonlighting functions
N.A.
Localizations
Stress granucle
BulkPerturb-seq
Ensembl ID ENSG00000143228 Gene ID
83540 Accession
14621
Symbol
NUF2
Alias
CDCA1;CT106;NUF2R
Full Name
NUF2 component of NDC80 kinetochore complex
Status
Confidence
Length
89189 bases
Strand
Plus strand
Position
1 : 163266576 - 163355764
RNA binding domain
N.A.
Summary
This gene encodes a protein that is highly similar to yeast Nuf2, a component of a conserved protein complex associated with the centromere. Yeast Nuf2 disappears from the centromere during meiotic prophase when centromeres lose their connection to the spindle pole body, and plays a regulatory role in chromosome segregation. The encoded protein is found to be associated with centromeres of mitotic HeLa cells, which suggests that this protein is a functional homolog of yeast Nuf2. Alternatively spliced transcript variants that encode the same protein have been described. [provided by RefSeq, Jul 2008]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
Name
Transcript ID
bp
Protein
Translation ID
NUF2-212
ENST00000534289
567
124aa
ENSP00000433533
NUF2-204
ENST00000450453
861
145aa
ENSP00000403252
NUF2-208
ENST00000524800
1845
417aa
ENSP00000436888
NUF2-203
ENST00000442820
1133
202aa
ENSP00000392652
NUF2-202
ENST00000367900
1842
464aa
ENSP00000356875
NUF2-201
ENST00000271452
1964
464aa
ENSP00000271452
NUF2-206
ENST00000490881
695
No protein
-
NUF2-205
ENST00000487578
779
No protein
-
NUF2-207
ENST00000497990
885
221aa
ENSP00000436473
NUF2-210
ENST00000527439
940
No protein
-
NUF2-209
ENST00000527120
351
No protein
-
NUF2-211
ENST00000531529
255
No protein
-
Pathway ID
Pathway Name
Source
ensgID
Trait
pValue
Pubmed ID
382 Respiratory Function Tests 3.08634497827468E-5 17903307 4359 Cholesterol, LDL 3.04599638960568E-5 17903299 40820 Platelet Function Tests 9.2544658E-006 -
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
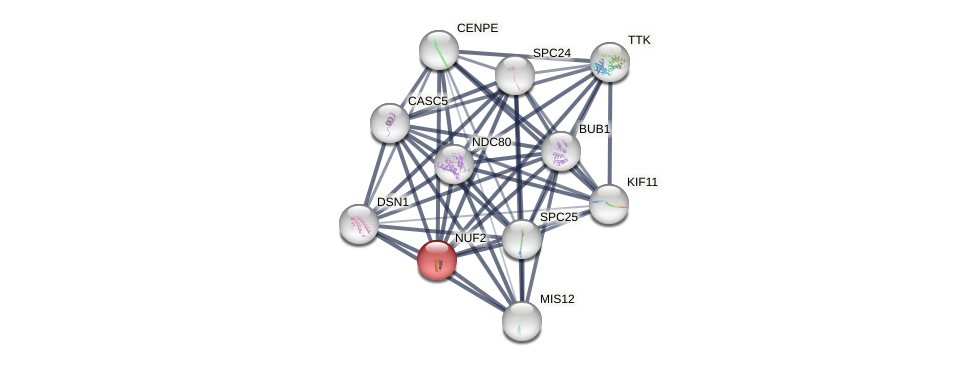
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0000775 located_in chromosome, centromeric region 1 NAS Homo_sapiens(9606) Component GO:0000776 located_in kinetochore 1 IDA Homo_sapiens(9606) Component GO:0003674 enables molecular_function 1 ND Homo_sapiens(9606) Function GO:0005515 enables protein binding 1 IPI Homo_sapiens(9606) Function GO:0005654 located_in nucleoplasm 1 IDA Homo_sapiens(9606) Component GO:0005829 located_in cytosol 2 IDA,TAS Homo_sapiens(9606) Component GO:0007052 involved_in mitotic spindle organization 1 IBA Homo_sapiens(9606) Process GO:0007059 involved_in chromosome segregation 1 NAS Homo_sapiens(9606) Process GO:0007094 involved_in mitotic spindle assembly checkpoint signaling 1 NAS Homo_sapiens(9606) Process GO:0008608 involved_in attachment of spindle microtubules to kinetochore 1 IDA Homo_sapiens(9606) Process GO:0016020 located_in membrane 1 HDA Homo_sapiens(9606) Component GO:0031262 part_of Ndc80 complex 3 IBA,IDA,IPI Homo_sapiens(9606) Component GO:0044877 enables protein-containing complex binding 1 IBA Homo_sapiens(9606) Function GO:0045132 involved_in meiotic chromosome segregation 1 IBA Homo_sapiens(9606) Process GO:0051301 involved_in cell division 1 IEA Homo_sapiens(9606) Process GO:0051315 involved_in attachment of mitotic spindle microtubules to kinetochore 1 IBA Homo_sapiens(9606) Process GO:0051383 involved_in kinetochore organization 1 IBA Homo_sapiens(9606) Process
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.