Welcome to
RBP World!
Functions and Diseases
| RBP Type | Canonical_RBPs |
| Diseases | Cancer |
| Drug | N.A. |
| Main interacting RNAs | rRNA |
| Moonlighting functions | E3 ligaseTransmemebrane |
| Localizations | Nucleoli |
| BulkPerturb-seq | N.A. |
Description
| Ensembl ID | ENSG00000135372 | Gene ID | 55226 | Accession | 29830 |
| Symbol | NAT10 | Alias | ALP;Kre33;NET43 | Full Name | N-acetyltransferase 10 |
| Status | Confidence | Length | 41292 bases | Strand | Plus strand |
| Position | 11 : 34105617 - 34146908 | RNA binding domain | tRNA_bind_2 | ||
| Summary | The protein encoded by this gene is an RNA cytidine acetyltransferase involved in histone acetylation, tRNA acetylation, the biosynthesis of 18S rRNA, and the enhancement of nuclear architecture and chromatin organization. [provided by RefSeq, Oct 2016] | ||||
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|---|---|---|---|
| 37484809 | NAT10-mediated RNA acetylation enhances HNRNPUL1 mRNA stability to contribute cervical cancer progression. | Yingfei Long | 2023-01-01 | International journal of medical sciences |
| 35721510 | The Key Role of RNA Modification in Breast Cancer. | Yang Liu | 2022-01-01 | Frontiers in cell and developmental biology |
| 30449621 | Acetylation of Cytidine in mRNA Promotes Translation Efficiency. | Daniel Arango | 2018-12-13 | Cell |
| 36765042 | Role of NAT10-mediated ac4C-modified HSP90AA1 RNA acetylation in ER stress-mediated metastasis and lenvatinib resistance in hepatocellular carcinoma. | Zhipeng Pan | 2023-02-10 | Cell death discovery |
| 37328448 | N4-Acetylcytidine Drives Glycolysis Addiction in Gastric Cancer via NAT10/SEPT9/HIF-1α Positive Feedback Loop. | Qingbin Yang | 2023-08-01 | Advanced science (Weinheim, Baden-Wurttemberg, Germany) |
| 37484432 | ac4C acetylation regulates mRNA stability and translation efficiency in osteosarcoma. | Wenjie Zhang | 2023-06-01 | Heliyon |
| 35971620 | N4-acetylcytidine regulates the replication and pathogenicity of enterovirus 71. | Haojie Hao | 2022-09-09 | Nucleic acids research |
| 37128278 | Mechanisms of NAT10 as ac4C writer in diseases. | Lihua Xie | 2023-06-13 | Molecular therapy. Nucleic acids |
| 36522719 | Acetyltransferase NAT10 regulates the Wnt/β-catenin signaling pathway to promote colorectal cancer progression via ac4C acetylation of KIF23 mRNA. | Chi Jin | 2022-12-15 | Journal of experimental & clinical cancer research : CR |
| 34362389 | NAT10 as a potential prognostic biomarker and therapeutic target for HNSCC. | Wenjie Tao | 2021-08-06 | Cancer cell international |
| 35978332 | The LINC00623/NAT10 signaling axis promotes pancreatic cancer progression by remodeling ac4C modification of mRNA. | Zengyu Feng | 2022-08-17 | Journal of hematology & oncology |
| 36209353 | N-acetyltransferase 10 promotes colon cancer progression by inhibiting ferroptosis through N4-acetylation and stabilization of ferroptosis suppressor protein 1 (FSP1) mRNA. | Xiao Zheng | 2022-12-01 | Cancer communications (London, England) |
| 35852833 | NAT10 and DDX21 Proteins Interact with RNase H1 and Affect the Performance of Phosphorothioate Oligonucleotides. | Lingdi Zhang | 2022-08-01 | Nucleic acid therapeutics |
| 36363698 | Learning from the Invaders: What Viruses Teach Us about RNA-Based Regulation in Microbes. | Peter L Sarin | 2022-10-25 | Microorganisms |
| 29970603 | Inhibition of the acetyltransferase NAT10 normalizes progeric and aging cells by rebalancing the Transportin-1 nuclear import pathway. | Delphine Larrieu | 2018-07-03 | Science signaling |
| 36949954 | Immune response and drug therapy based on ac4C-modified gene in pancreatic cancer typing. | Dong Xu | 2023-01-01 | Frontiers in immunology |
| 34395433 | NAT10-Mediated N4-Acetylcytidine of RNA Contributes to Post-transcriptional Regulation of Mouse Oocyte Maturation in vitro. | Yuting Xiang | 2021-01-01 | Frontiers in cell and developmental biology |
| 36908042 | NAT10 mediated mRNA acetylation modification patterns associated with colon cancer progression and microsatellite status. | Hailin Zhang | 2023-12-01 | Epigenetics |
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| NAT10-207 | ENST00000532503 | 556 | No protein | - |
| NAT10-205 | ENST00000531159 | 3436 | 953aa | ENSP00000433011 |
| NAT10-201 | ENST00000257829 | 3973 | 1025aa | ENSP00000257829 |
| NAT10-206 | ENST00000531723 | 528 | No protein | - |
| NAT10-203 | ENST00000529523 | 561 | 145aa | ENSP00000435569 |
| NAT10-202 | ENST00000527971 | 1600 | 288aa | ENSP00000437324 |
| NAT10-204 | ENST00000530017 | 543 | No protein | - |
| NAT10-208 | ENST00000532555 | 615 | No protein | - |
| NAT10-209 | ENST00000534509 | 420 | No protein | - |
Phenotypes
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| 26470 | Blood Pressure | 7.8870000E-005 | - |
| 60302 | Blood Pressure | 1.6594968E-005 | - |
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
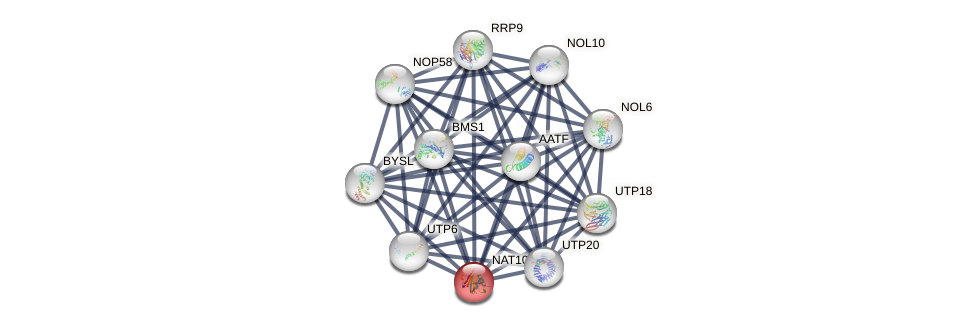
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000135372 | ||||||||
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 99.9024 | 99.2195 | nomascus_leucogenys | ENSNLEP00000021769 | 99.9024 | 99.2195 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 99.0244 | 98.3415 | pan_paniscus | ENSPPAP00000025919 | 98.1625 | 97.4855 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 99.8049 | 99.2195 | pongo_abelii | ENSPPYP00000003867 | 99.8049 | 99.2195 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 98.9268 | 98.2439 | pan_troglodytes | ENSPTRP00000006068 | 98.0658 | 97.3888 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 99.0244 | 98.1463 | gorilla_gorilla | ENSGGOP00000034321 | 98.1625 | 97.2921 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 93.2683 | 87.5122 | ornithorhynchus_anatinus | ENSOANP00000011778 | 96.5657 | 90.6061 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 95.7073 | 88.4878 | sarcophilus_harrisii | ENSSHAP00000003911 | 95.614 | 88.4016 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 66.6341 | 62.5366 | notamacropus_eugenii | ENSMEUP00000011615 | 71.6684 | 67.2613 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 78.2439 | 74.9268 | choloepus_hoffmanni | ENSCHOP00000001445 | 78.3203 | 75 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 98.5366 | 95.6098 | dasypus_novemcinctus | ENSDNOP00000029240 | 97.9631 | 95.0533 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 88.5854 | 86.2439 | erinaceus_europaeus | ENSEEUP00000006361 | 88.7586 | 86.4125 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 59.6098 | 55.8049 | echinops_telfairi | ENSETEP00000016274 | 62.0305 | 58.0711 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 99.2195 | 98.0488 | callithrix_jacchus | ENSCJAP00000087933 | 98.2609 | 97.1014 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 99.8049 | 99.0244 | cercocebus_atys | ENSCATP00000029134 | 99.7076 | 98.9279 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 95.7073 | 94.9268 | macaca_fascicularis | ENSMFAP00000007873 | 99.6951 | 98.8821 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 99.8049 | 99.1219 | macaca_mulatta | ENSMMUP00000064991 | 99.7076 | 99.0253 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 99.7073 | 98.8293 | macaca_nemestrina | ENSMNEP00000036301 | 99.6101 | 98.7329 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 99.9024 | 99.3171 | papio_anubis | ENSPANP00000001919 | 99.8051 | 99.2203 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 95.8049 | 95.3171 | mandrillus_leucophaeus | ENSMLEP00000003900 | 99.4934 | 98.9868 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 84.1951 | 82.2439 | tursiops_truncatus | ENSTTRP00000007332 | 84.2773 | 82.3242 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 95.1219 | 92.6829 | vulpes_vulpes | ENSVVUP00000013925 | 98.4848 | 95.9596 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 97.9512 | 95.0244 | mus_spretus | MGP_SPRETEiJ_P0055485 | 98.0469 | 95.1172 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 97.6585 | 95.1219 | equus_asinus | ENSEASP00005029729 | 97.5634 | 95.0292 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 98.7317 | 96.5854 | capra_hircus | ENSCHIP00000031917 | 98.6355 | 96.4912 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 99.0244 | 95.7073 | loxodonta_africana | ENSLAFP00000010367 | 98.7354 | 95.428 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 98.1463 | 96.0976 | panthera_leo | ENSPLOP00000008809 | 98.0507 | 96.0039 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 98.7317 | 95.8049 | ailuropoda_melanoleuca | ENSAMEP00000039507 | 98.6355 | 95.7115 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 98.5366 | 96.6829 | bos_indicus_hybrid | ENSBIXP00005033811 | 98.4405 | 96.5887 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 96.2927 | 93.6585 | oryctolagus_cuniculus | ENSOCUP00000001205 | 95.8252 | 93.2039 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 97.4634 | 95.0244 | equus_caballus | ENSECAP00000028158 | 91.9043 | 89.6044 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 98.439 | 95.8049 | canis_lupus_familiaris | ENSCAFP00845033715 | 98.439 | 95.8049 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 98.1463 | 96.0976 | panthera_pardus | ENSPPRP00000018588 | 98.0507 | 96.0039 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 98.3415 | 96.2927 | marmota_marmota_marmota | ENSMMMP00000011257 | 98.2456 | 96.1988 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 97.7561 | 94.8293 | balaenoptera_musculus | ENSBMSP00010018104 | 97.8516 | 94.9219 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 97.6585 | 93.9512 | cavia_porcellus | ENSCPOP00000030709 | 97.1845 | 93.4951 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 98.3415 | 96.2927 | felis_catus | ENSFCAP00000001875 | 98.2456 | 96.1988 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 97.3659 | 93.6585 | ursus_americanus | ENSUAMP00000013871 | 96.5184 | 92.8433 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 95.6098 | 88.3902 | monodelphis_domestica | ENSMODP00000012382 | 95.4236 | 88.2181 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 98.3415 | 96.2927 | bison_bison_bison | ENSBBBP00000002973 | 98.2456 | 96.1988 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 97.7561 | 94.5366 | monodon_monoceros | ENSMMNP00015027442 | 97.6608 | 94.4444 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 98.6341 | 96.2927 | sus_scrofa | ENSSSCP00000014137 | 98.538 | 96.1988 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 97.0732 | 95.4146 | microcebus_murinus | ENSMICP00000003324 | 97.3581 | 95.6947 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 97.7561 | 94.9268 | octodon_degus | ENSODEP00000023286 | 97.7561 | 94.9268 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 98.439 | 95.8049 | jaculus_jaculus | ENSJJAP00000023210 | 98.3431 | 95.7115 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 98.6341 | 96.6829 | ovis_aries_rambouillet | ENSOARP00020014608 | 98.538 | 96.5887 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 98.7317 | 95.4146 | ursus_maritimus | ENSUMAP00000024914 | 98.6355 | 95.3216 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 93.2683 | 90.8293 | ochotona_princeps | ENSOPRP00000008940 | 96.7611 | 94.2308 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 97.7561 | 94.7317 | physeter_catodon | ENSPCTP00005014582 | 97.6608 | 94.6394 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 18.1463 | 17.6585 | mus_caroli | MGP_CAROLIEiJ_P0053407 | 100 | 97.3118 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 98.1463 | 95.4146 | mus_musculus | ENSMUSP00000028608 | 98.2422 | 95.5078 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 97.9512 | 95.1219 | dipodomys_ordii | ENSDORP00000006153 | 97.7605 | 94.9367 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 98.439 | 95.4146 | mustela_putorius_furo | ENSMPUP00000011544 | 98.3431 | 95.3216 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 99.5122 | 98.439 | aotus_nancymaae | ENSANAP00000018044 | 99.4152 | 98.3431 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 99.5122 | 98.3415 | saimiri_boliviensis_boliviensis | ENSSBOP00000013782 | 99.4152 | 98.2456 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 99.5122 | 98.7317 | chlorocebus_sabaeus | ENSCSAP00000001695 | 99.4152 | 98.6355 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 66.5366 | 61.9512 | tupaia_belangeri | ENSTBEP00000008943 | 68.9585 | 64.2063 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 76.8781 | 74.3415 | mesocricetus_auratus | ENSMAUP00000009549 | 96.925 | 93.7269 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 18.1463 | 17.7561 | mesocricetus_auratus | ENSMAUP00000008641 | 100 | 97.8495 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 97.561 | 94.3415 | phocoena_sinus | ENSPSNP00000024065 | 97.4659 | 94.2495 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 98.8293 | 96.2927 | camelus_dromedarius | ENSCDRP00005025953 | 98.7329 | 96.1988 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 95.9024 | 89.4634 | carlito_syrichta | ENSTSYP00000012405 | 95.809 | 89.3762 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 98.1463 | 96.0976 | panthera_tigris_altaica | ENSPTIP00000009727 | 98.0507 | 96.0039 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 97.8537 | 95.0244 | rattus_norvegicus | ENSRNOP00000012094 | 97.9492 | 95.1172 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 98.3415 | 96.8781 | prolemur_simus | ENSPSMP00000023154 | 98.2456 | 96.7836 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 97.7561 | 95.0244 | microtus_ochrogaster | ENSMOCP00000025296 | 97.7561 | 95.0244 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 61.0732 | 59.8049 | sorex_araneus | ENSSARP00000012184 | 78.5445 | 76.9134 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 97.9512 | 95.1219 | peromyscus_maniculatus_bairdii | ENSPEMP00000007531 | 97.9512 | 95.1219 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 97.8537 | 96 | bos_mutus | ENSBMUP00000011186 | 97.7583 | 95.9064 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 98.0488 | 95.2195 | mus_spicilegus | ENSMSIP00000007375 | 98.1445 | 95.3125 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 98.0488 | 95.4146 | cricetulus_griseus_chok1gshd | ENSCGRP00001008249 | 98.0488 | 95.4146 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 89.4634 | 87.8049 | pteropus_vampyrus | ENSPVAP00000005267 | 89.5508 | 87.8906 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 97.7561 | 95.2195 | myotis_lucifugus | ENSMLUP00000015019 | 97.4708 | 94.9416 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 95.5122 | 88.4878 | vombatus_ursinus | ENSVURP00010016476 | 95.4191 | 88.4016 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 99.0244 | 96.3902 | rhinolophus_ferrumequinum | ENSRFEP00010007447 | 98.9279 | 96.2963 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 98.3415 | 95.2195 | neovison_vison | ENSNVIP00000014837 | 98.2456 | 95.1267 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 98.439 | 96.4878 | bos_taurus | ENSBTAP00000022271 | 98.439 | 96.4878 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 97.6585 | 93.9512 | heterocephalus_glaber_female | ENSHGLP00000023674 | 97.5634 | 93.8596 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 99.8049 | 98.8293 | rhinopithecus_bieti | ENSRBIP00000010137 | 99.7076 | 98.7329 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 95.1219 | 92.8781 | canis_lupus_dingo | ENSCAFP00020005554 | 98.4848 | 96.1616 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 98.7317 | 96.3902 | sciurus_vulgaris | ENSSVLP00005006041 | 98.6355 | 96.2963 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 96.2927 | 89.2683 | phascolarctos_cinereus | ENSPCIP00000053931 | 93.4659 | 86.6477 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 81.8537 | 78.0488 | procavia_capensis | ENSPCAP00000004121 | 84.8332 | 80.8898 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 97.8537 | 95.2195 | nannospalax_galili | ENSNGAP00000011312 | 97.7583 | 95.1267 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 98.5366 | 96.6829 | bos_grunniens | ENSBGRP00000030600 | 98.4405 | 96.5887 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 97.561 | 94.5366 | chinchilla_lanigera | ENSCLAP00000024312 | 97.4659 | 94.4444 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 98.439 | 96.3902 | cervus_hanglu_yarkandensis | ENSCHYP00000015950 | 98.3431 | 96.2963 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 98.439 | 95.4146 | catagonus_wagneri | ENSCWAP00000029170 | 98.3431 | 95.3216 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 98.439 | 96.4878 | ictidomys_tridecemlineatus | ENSSTOP00000001635 | 98.3431 | 96.3938 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 22.6341 | 20.4878 | ictidomys_tridecemlineatus | ENSSTOP00000028374 | 85.6089 | 77.4908 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 98.6341 | 97.1707 | otolemur_garnettii | ENSOGAP00000014983 | 98.6341 | 97.1707 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 99.5122 | 98.2439 | cebus_imitator | ENSCCAP00000000855 | 99.4152 | 98.1481 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 98.7317 | 96.6829 | urocitellus_parryii | ENSUPAP00010024537 | 98.6355 | 96.5887 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 98.439 | 96.4878 | moschus_moschiferus | ENSMMSP00000017866 | 98.3431 | 96.3938 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 99.8049 | 98.9268 | rhinopithecus_roxellana | ENSRROP00000002952 | 99.7076 | 98.8304 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 98.1463 | 94.9268 | mus_pahari | MGP_PahariEiJ_P0064371 | 98.2422 | 95.0195 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 98.7317 | 97.1707 | propithecus_coquereli | ENSPCOP00000003186 | 98.6355 | 97.076 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 72.0976 | 54.5366 | caenorhabditis_elegans | F55A12.8.1 | 70.8533 | 53.5954 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 69.4634 | 52.2927 | drosophila_melanogaster | FBpp0071217 | 70.6349 | 53.1746 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 76.1951 | 60.9756 | ciona_intestinalis | ENSCINP00000007371 | 81.6946 | 65.3766 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 89.0732 | 77.4634 | petromyzon_marinus | ENSPMAP00000004016 | 88.9864 | 77.3879 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 87.4146 | 75.1219 | eptatretus_burgeri | ENSEBUP00000020420 | 87.6712 | 75.3425 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 91.2195 | 80.2927 | callorhinchus_milii | ENSCMIP00000037865 | 85.6227 | 75.3663 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 91.7073 | 82.0488 | latimeria_chalumnae | ENSLACP00000005218 | 91.7073 | 82.0488 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 89.2683 | 80.5854 | lepisosteus_oculatus | ENSLOCP00000003423 | 91.3174 | 82.4351 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 87.1219 | 76.5854 | clupea_harengus | ENSCHAP00000013675 | 86.0308 | 75.6262 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 90.0488 | 80.3902 | danio_rerio | ENSDARP00000070873 | 90.0488 | 80.3902 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 90.5366 | 80.1951 | carassius_auratus | ENSCARP00000129744 | 89.7486 | 79.4971 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 90.439 | 81.3659 | astyanax_mexicanus | ENSAMXP00000028570 | 90.7045 | 81.6047 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 25.0732 | 22.3415 | ictalurus_punctatus | ENSIPUP00000010240 | 91.1348 | 81.2057 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 89.7561 | 80.2927 | esox_lucius | ENSELUP00000004833 | 89.061 | 79.6709 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 90.7317 | 80.9756 | electrophorus_electricus | ENSEEEP00000022126 | 87.9849 | 78.5241 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 89.1707 | 79.5122 | oncorhynchus_kisutch | ENSOKIP00005024211 | 87.1306 | 77.693 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 90.3415 | 80.1951 | oncorhynchus_mykiss | ENSOMYP00000104620 | 89.3822 | 79.3436 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 90.2439 | 80.1951 | salmo_salar | ENSSSAP00000173358 | 85.7275 | 76.1816 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 88.8781 | 79.2195 | salmo_trutta | ENSSTUP00000092577 | 85.9434 | 76.6038 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 88.5854 | 76.9756 | gadus_morhua | ENSGMOP00000034265 | 88.1553 | 76.6019 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 90.5366 | 79.7073 | fundulus_heteroclitus | ENSFHEP00000026331 | 89.6618 | 78.9372 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 83.5122 | 73.6585 | poecilia_reticulata | ENSPREP00000023915 | 89.6335 | 79.0576 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 90.439 | 79.4146 | xiphophorus_maculatus | ENSXMAP00000036876 | 89.8256 | 78.876 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 89.8537 | 79.3171 | oryzias_latipes | ENSORLP00000006083 | 88.8139 | 78.3992 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 89.6585 | 79.6098 | cyclopterus_lumpus | ENSCLMP00005025108 | 88.621 | 78.6885 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 89.561 | 79.5122 | oreochromis_niloticus | ENSONIP00000035147 | 87.0142 | 77.2512 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 89.561 | 79.4146 | haplochromis_burtoni | ENSHBUP00000002504 | 88.6957 | 78.6473 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 89.6585 | 79.5122 | astatotilapia_calliptera | ENSACLP00000010812 | 88.7923 | 78.744 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 89.7561 | 79.8049 | sparus_aurata | ENSSAUP00010043786 | 88.7175 | 78.8814 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 90.6341 | 80.7805 | lates_calcarifer | ENSLCAP00010017017 | 89.499 | 79.7688 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 90.7317 | 80.9756 | xenopus_tropicalis | ENSXETP00000008215 | 93.9394 | 83.8384 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 92.0976 | 85.2683 | chrysemys_picta_bellii | ENSCPBP00000017041 | 92.3679 | 85.5186 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 7.31707 | 6.82927 | sphenodon_punctatus | ENSSPUP00000000258 | 92.5926 | 86.4198 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 93.2683 | 85.9512 | sphenodon_punctatus | ENSSPUP00000013873 | 93.8175 | 86.4573 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 94.0488 | 86.1463 | crocodylus_porosus | ENSCPRP00005015337 | 91.8095 | 84.0952 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 55.5122 | 52 | laticauda_laticaudata | ENSLLTP00000007579 | 90.7496 | 85.008 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 57.2683 | 51.5122 | notechis_scutatus | ENSNSUP00000012460 | 83.3807 | 75 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 92.2927 | 84.8781 | pseudonaja_textilis | ENSPTXP00000010376 | 92.8361 | 85.3778 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 94.6341 | 87.0244 | anas_platyrhynchos_platyrhynchos | ENSAPLP00000007357 | 95.1914 | 87.5368 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 94.3415 | 87.1219 | gallus_gallus | ENSGALP00010037513 | 94.897 | 87.6349 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 92.9756 | 85.3659 | meleagris_gallopavo | ENSMGAP00000010594 | 88.9823 | 81.6993 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 94.0488 | 87.0244 | serinus_canaria | ENSSCAP00000011900 | 94.6026 | 87.5368 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 94.1463 | 87.0244 | parus_major | ENSPMJP00000009349 | 83.913 | 77.5652 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 92.7805 | 85.561 | anolis_carolinensis | ENSACAP00000013354 | 93.3268 | 86.0648 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 89.3659 | 79.3171 | neolamprologus_brichardi | ENSNBRP00000002901 | 89.2788 | 79.2398 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 82.5366 | 74.8293 | naja_naja | ENSNNAP00000011074 | 91.1638 | 82.6509 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 91.6098 | 84.6829 | podarcis_muralis | ENSPMRP00000003147 | 89.3435 | 82.588 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 87.7073 | 80.5854 | geospiza_fortis | ENSGFOP00000012804 | 88.9219 | 81.7013 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 93.9512 | 86.8293 | taeniopygia_guttata | ENSTGUP00000035068 | 94.1349 | 86.999 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 92.6829 | 82.3415 | erpetoichthys_calabaricus | ENSECRP00000004826 | 92.8641 | 82.5024 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 90.439 | 79.7073 | kryptolebias_marmoratus | ENSKMAP00000001900 | 89.3925 | 78.785 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 93.3659 | 86.1463 | salvator_merianae | ENSSMRP00000005233 | 94.0079 | 86.7387 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 90.3415 | 78.9268 | oryzias_javanicus | ENSOJAP00000015589 | 84.8763 | 74.1522 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 86.6341 | 76.2927 | amphilophus_citrinellus | ENSACIP00000022987 | 87.0588 | 76.6667 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 89.8537 | 79.9024 | dicentrarchus_labrax | ENSDLAP00005034297 | 88.8139 | 78.9778 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 90.439 | 80.7805 | amphiprion_percula | ENSAPEP00000001608 | 89.3925 | 79.8457 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 89.8537 | 79.1219 | oryzias_sinensis | ENSOSIP00000012282 | 88.0497 | 77.5335 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 92.2927 | 84.6829 | pelodiscus_sinensis | ENSPSIP00000007208 | 92.5636 | 84.9315 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 92.8781 | 85.4634 | gopherus_evgoodei | ENSGEVP00005023388 | 93.2419 | 85.7982 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 93.2683 | 86.3415 | chelonoidis_abingdonii | ENSCABP00000029723 | 90.4447 | 83.7275 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 90.0488 | 80.0976 | seriola_dumerili | ENSSDUP00000030389 | 89.0068 | 79.1707 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 89.8537 | 79.8049 | cottoperca_gobio | ENSCGOP00000051459 | 88.8139 | 78.8814 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 93.3659 | 85.6585 | struthio_camelus_australis | ENSSCUP00000025643 | 92.4638 | 84.8309 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 92.9756 | 86.1463 | anser_brachyrhynchus | ENSABRP00000020926 | 94.077 | 87.1668 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 89.2683 | 79.3171 | gasterosteus_aculeatus | ENSGACP00000013022 | 88.3205 | 78.4749 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 90.0488 | 79.6098 | cynoglossus_semilaevis | ENSCSEP00000020563 | 88.921 | 78.6127 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 90.1463 | 79.8049 | poecilia_formosa | ENSPFOP00000012930 | 89.4482 | 79.1868 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 89.8537 | 80.4878 | myripristis_murdjan | ENSMMDP00005001655 | 87.4644 | 78.3476 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 89.561 | 80.0976 | sander_lucioperca | ENSSLUP00000003285 | 88.5246 | 79.1707 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 94.6341 | 86.3415 | strigops_habroptila | ENSSHBP00005001969 | 95.1914 | 86.8499 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 90.439 | 80.7805 | amphiprion_ocellaris | ENSAOCP00000020426 | 89.3925 | 79.8457 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 93.6585 | 86.8293 | terrapene_carolina_triunguis | ENSTMTP00000011716 | 94.21 | 87.3405 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 90.3415 | 80.3902 | hucho_hucho | ENSHHUP00000035251 | 89.4686 | 79.6135 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 89.2683 | 78.6341 | betta_splendens | ENSBSLP00000037719 | 88.2353 | 77.7242 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 89.3659 | 80.3902 | pygocentrus_nattereri | ENSPNAP00000013381 | 89.4531 | 80.4688 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 89.1707 | 79.5122 | anabas_testudineus | ENSATEP00000049406 | 86.2264 | 76.8868 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 73.7561 | 57.1707 | saccharomyces_cerevisiae | YNL132W | 71.5909 | 55.4924 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 90.0488 | 80 | seriola_lalandi_dorsalis | ENSSLDP00000006697 | 89.0068 | 79.0742 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 90.439 | 79.8049 | labrus_bergylta | ENSLBEP00000017232 | 89.2204 | 78.7295 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 89.4634 | 79.4146 | pundamilia_nyererei | ENSPNYP00000029042 | 88.599 | 78.6473 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 87.9024 | 78.7317 | mastacembelus_armatus | ENSMAMP00000000511 | 85.8095 | 76.8571 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 89.9512 | 80.7805 | stegastes_partitus | ENSSPAP00000018426 | 88.9103 | 79.8457 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 90.3415 | 80.4878 | oncorhynchus_tshawytscha | ENSOTSP00005045025 | 87.2762 | 77.7568 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 90.3415 | 80.4878 | oncorhynchus_tshawytscha | ENSOTSP00005048288 | 86.2197 | 76.8156 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 90.5366 | 80.7805 | acanthochromis_polyacanthus | ENSAPOP00000008436 | 89.4889 | 79.8457 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 94.2439 | 86.9268 | aquila_chrysaetos_chrysaetos | ENSACCP00020020872 | 94.7988 | 87.4387 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 90.1463 | 79.4146 | nothobranchius_furzeri | ENSNFUP00015045988 | 88.8462 | 78.2692 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 24.7805 | 21.1707 | cyprinodon_variegatus | ENSCVAP00000018707 | 90.0709 | 76.9504 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 89.8537 | 80.2927 | denticeps_clupeoides | ENSDCDP00000057816 | 89.9414 | 80.3711 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 89.7561 | 79.8049 | larimichthys_crocea | ENSLCRP00005054832 | 88.7175 | 78.8814 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 25.9512 | 23.122 | takifugu_rubripes | ENSTRUP00000043718 | 90.4762 | 80.6122 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 93.9512 | 86.9268 | ficedula_albicollis | ENSFALP00000001338 | 91.8017 | 84.938 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 89.4634 | 78.7317 | hippocampus_comes | ENSHCOP00000026121 | 88.599 | 77.971 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 90.5366 | 80.8781 | cyprinus_carpio_carpio | ENSCCRP00000035753 | 90.3603 | 80.7205 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 90.439 | 81.0732 | cyprinus_carpio_carpio | ENSCCRP00000035823 | 90.439 | 81.0732 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 93.0732 | 86.0488 | coturnix_japonica | ENSCJPP00005019159 | 93.0732 | 86.0488 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 90.1463 | 79.8049 | poecilia_latipinna | ENSPLAP00000011284 | 89.4482 | 79.1868 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 90.1463 | 80.0976 | sinocyclocheilus_grahami | ENSSGRP00000044492 | 89.9708 | 79.9416 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 89.9512 | 80.1951 | sinocyclocheilus_grahami | ENSSGRP00000076779 | 88.0611 | 78.51 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 89.561 | 79.4146 | maylandia_zebra | ENSMZEP00005006520 | 88.6957 | 78.6473 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 90.2439 | 80.2927 | tetraodon_nigroviridis | ENSTNIP00000019133 | 89.1996 | 79.3635 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 90.7317 | 81.6585 | paramormyrops_kingsleyae | ENSPKIP00000018623 | 90.9091 | 81.8182 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 91.0244 | 82.439 | scleropages_formosus | ENSSFOP00015055453 | 89.2823 | 80.8612 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 87.9024 | 78.5366 | leptobrachium_leishanense | ENSLLEP00000024972 | 92.3156 | 82.4795 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 89.9512 | 80 | scophthalmus_maximus | ENSSMAP00000054213 | 84.8206 | 75.437 |
| ENSG00000135372 | homo_sapiens | ENSP00000257829 | 90.1463 | 79.3171 | oryzias_melastigma | ENSOMEP00000012457 | 84.7706 | 74.5872 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0000049 | enables tRNA binding | 1 | IBA | Homo_sapiens(9606) | Function |
| GO:0000154 | involved_in rRNA modification | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0000781 | located_in chromosome, telomeric region | 1 | HDA | Homo_sapiens(9606) | Component |
| GO:0003723 | enables RNA binding | 1 | HDA | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005524 | enables ATP binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0005634 | located_in nucleus | 1 | NAS | Homo_sapiens(9606) | Component |
| GO:0005654 | located_in nucleoplasm | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0005697 | part_of telomerase holoenzyme complex | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005730 | is_active_in nucleolus | 2 | IBA,IDA | Homo_sapiens(9606) | Component |
| GO:0006473 | involved_in protein acetylation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0008080 | enables N-acetyltransferase activity | 1 | EXP | Homo_sapiens(9606) | Function |
| GO:0010824 | involved_in regulation of centrosome duplication | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0016020 | located_in membrane | 1 | HDA | Homo_sapiens(9606) | Component |
| GO:0030496 | located_in midbody | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0032040 | part_of small-subunit processome | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0032211 | involved_in negative regulation of telomere maintenance via telomerase | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0042274 | involved_in ribosomal small subunit biogenesis | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0045727 | involved_in positive regulation of translation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0051391 | involved_in tRNA acetylation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0070182 | enables DNA polymerase binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0106162 | enables mRNA N-acetyltransferase activity | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:1904812 | involved_in rRNA acetylation involved in maturation of SSU-rRNA | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:1990883 | enables rRNA cytidine N-acetyltransferase activity | 1 | IBA | Homo_sapiens(9606) | Function |